Chromatin region accessibility: cross-species comparisons and cell type distinctions
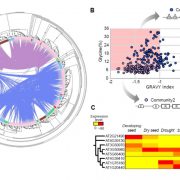
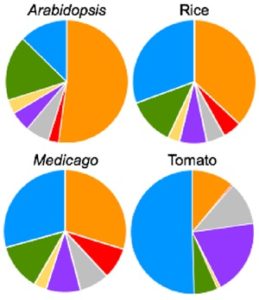 Cis-regulatory elements direct transcriptional output during cell differentiation and coordinate adaptive responses to environmental stress. DNA becomes open and accessible when nucleosomes are displaced by DNA-binding proteins, indicating a correlation between open chromatin regions and transcriptional regulation. Utilizing four different plant species and several distinct methods (i.e. INTACT for nuclei isolation and ATAC-seq for open chromatin identification), Maher et al. investigated cis-regulatory elements in accessible chromatin regions, (transposase hypersensitive sites or THSs), of isolated root tip nuclei. The overall proportion of THSs were similar in cross-species comparisons, with the majority of THSs being extragenic and within 3 kb upstream of a transcription state site. However, the number and location of open chromatin regions were highly variable. On average, genes tended to have only one THS, if any; orthologous genes displayed no conservation cross-species in the number or location of THSs; and THSs did not correlate with expression level. Extensive overlap was seen in THSs between distinct cell types, and THSs specific to a cell type, named differential THSs (dTHSs), were also found. Interestingly, dTHSs were found near abundantly- and minimally- or not-expressed genes. The authors demonstrated and compared multiple methods to explore chromatin accessibility in distinct cell types and constructed models of TF regulatory networks, providing valuable techniques to the field and a better understanding of the evolution of plant gene regulatory and cell fate specification mechanisms. (Summary by Alecia Biel) Plant Cell. 10.1105/tpc.17.00581.
Cis-regulatory elements direct transcriptional output during cell differentiation and coordinate adaptive responses to environmental stress. DNA becomes open and accessible when nucleosomes are displaced by DNA-binding proteins, indicating a correlation between open chromatin regions and transcriptional regulation. Utilizing four different plant species and several distinct methods (i.e. INTACT for nuclei isolation and ATAC-seq for open chromatin identification), Maher et al. investigated cis-regulatory elements in accessible chromatin regions, (transposase hypersensitive sites or THSs), of isolated root tip nuclei. The overall proportion of THSs were similar in cross-species comparisons, with the majority of THSs being extragenic and within 3 kb upstream of a transcription state site. However, the number and location of open chromatin regions were highly variable. On average, genes tended to have only one THS, if any; orthologous genes displayed no conservation cross-species in the number or location of THSs; and THSs did not correlate with expression level. Extensive overlap was seen in THSs between distinct cell types, and THSs specific to a cell type, named differential THSs (dTHSs), were also found. Interestingly, dTHSs were found near abundantly- and minimally- or not-expressed genes. The authors demonstrated and compared multiple methods to explore chromatin accessibility in distinct cell types and constructed models of TF regulatory networks, providing valuable techniques to the field and a better understanding of the evolution of plant gene regulatory and cell fate specification mechanisms. (Summary by Alecia Biel) Plant Cell. 10.1105/tpc.17.00581.