APE2 Acts in DNA Demethylation
Li et al. demonstrate that APE2 and the DNA 3′ phosphatase ZDP play overlapping roles in active DNA demethylation in Arabidopsis thaliana. The Plant Cell (2018).
By Jinchao Li and Weiqiang Qian
Background: DNA methylation, the addition of a methyl group to the C-5 position of cytosine, is a type of DNA modification important for gene expression regulation. DNA methylation is reversible. In eukaryotes, the active removal of methylated cytosine, referred to as active DNA demethylation, employs the base excision repair pathway, a pathway evolved for repair of damaged DNA bases. In plants, the ROS1 family of DNA glycosylases cut the DNA backbone after removing methylated cytosine, generating a gap with a 3′ phosphate or a 3′ phosphor-α,β-unsaturated aldehyde (PUA) terminus. ZDP and APE1L convert 3′ phosphate and 3′ PUA, respectively, to a 3′ hydroxyl group. The conversions enable gap filling with an unmethylated cytosine by DNA polymerase(s) and DNA ligase I.
Question: We wanted to know whether APE2, another apurinic/apyrimidinic endonuclease protein, functions in active DNA demethylation and DNA damage repair. What is the relationship between APE2 and ZDP in these biological processes?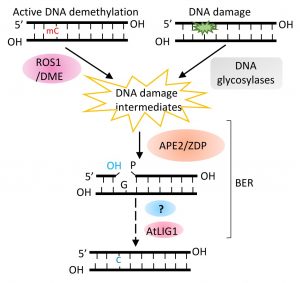
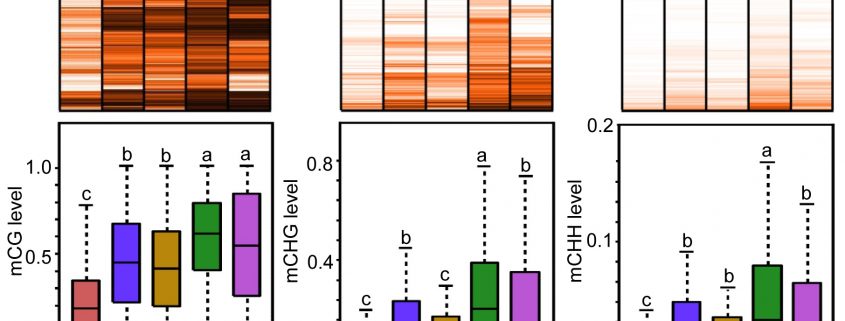
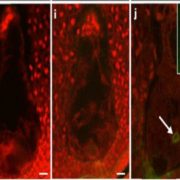
Findings: We found that APE2 functions in active DNA demethylation. Unexpectedly, APE2 and ZDP play overlapping roles, downstream of ROS1, in active DNA demethylation. The zdp ape2 double mutants, but not the single mutants, display many defects associated with DNA repair deficiency, suggesting overlapping roles of APE2 and ZDP in DNA damage repair as well. Knockout of ROS1 reduces the magnitude of DNA lesion accumulation and the DNA damage response in the zdp ape2 mutants, suggesting that ROS1 products, when kept unprocessed and unrepaired, are an important source of DNA damage. Finally, we demonstrate that APE2, like ZDP, has DNA 3′ phosphatase activity. Besides, it has strong 3′ to 5′ exonuclease activity.
Next steps: What mechanisms determine the specificity of APE2 and ZDP2 targeting to certain loci. What are the DNA damage signaling pathways, for instance, the checkpoint pathways, that are specifically activated in the zdp ape2 double mutants, which show defects in both genome and epigenome stability.
Jinchao Li, Wenjie Liang, Yan Li, and Weiqiang Qian (2018). APURINIC/APYRIMIDINIC ENDONUCLEASE2 and ZINC FINGER DNA 3′-PHOSPHOESTERASE Play Overlapping Roles in the Maintenance of Epigenome and Genome Stability. Plant Cell 30: 1954-1970; DOI: https://doi.org/10.1105/tpc.18.00287
Key words: DNA methylation; DNA damage; base excision repair; DNA damage response