Another Brick in the Plant Cell Wall: Characterization of Arabidopsis CSLD3 Function in Cell Wall Synthesis
Plant cell walls are one of the great engineering feats of nature, providing immense structural strength and durability. The core components of plant cell walls (pectin, hemicellulose, and cellulose) can occur in different proportions and vary in their structure and assembly, thus allowing cell walls to be tailored for specific purposes. To achieve this variability, construction of plant cell walls requires a complex set of enzymatic machinery. Our knowledge of this machinery, particularly the function of members of the expanded cellulose synthase (CESA) and cellulose synthase like (CSL) gene families, remains highly fragmented. However, continued research is revealing, brick by brick, how each enzyme contributes to building the components of cell walls.
 The function of members of the CSLD clade has been associated with the polarized cell wall deposition of tip growing cells across the plant kingdom. In both root hairs of Arabidopsis thaliana and rhizoids of Marchantia polymorpha, loss of CSLD isoforms severely inhibited elongation and led to cytoplasmic leakage, suggesting a compromised cell wall (Wang et al., 2001; Honkanen et al., 2016). However, it was unclear which type of polysaccharides members of the CSLD clade were producing. Now, Yang et al. (2020) have confirmed the biochemical role of one cell wall builder, CSLD3, by determining its catalytic activity as a β-1,4-glucan synthase.
The function of members of the CSLD clade has been associated with the polarized cell wall deposition of tip growing cells across the plant kingdom. In both root hairs of Arabidopsis thaliana and rhizoids of Marchantia polymorpha, loss of CSLD isoforms severely inhibited elongation and led to cytoplasmic leakage, suggesting a compromised cell wall (Wang et al., 2001; Honkanen et al., 2016). However, it was unclear which type of polysaccharides members of the CSLD clade were producing. Now, Yang et al. (2020) have confirmed the biochemical role of one cell wall builder, CSLD3, by determining its catalytic activity as a β-1,4-glucan synthase.
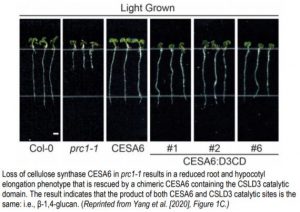
To characterize the function of Arabidopsis CSLD3, the authors synthesized chimeric versions of CSLD3 and Arabidopsis CESA6, which is a known cellulose synthase. Park et al. (2011) observed that expressing a chimeric CSLD3 protein containing the CESA6 catalytic domain rescued the csld3 tip growth phenotype in Arabidopsis. This suggested that the catalytic domain of CSLD3 mirrored that of CESA6, which produces β-1,4-glucan. Now, the reciprocal approach has been taken by creating a CESA6 chimera containing the CSLD3 catalytic domain (Yang et al., 2020). Expression of this chimeric protein under the endogenous CESA6 promoter rescued the cesa6 (prc1-1) root and hypocotyl elongation phenotype, which results from reduced cellulose content in Arabidopsis (Figure). Furthermore, use of a catalytically inactive CSLD3 domain could not rescue the cesa6 phenotype, confirming that the CSLD3 active site was responsible for the rescued cellulose deficiency phenotype.
The catalytic activity of CSLD3 was confirmed by in vitro enzyme assays using Arabidopsis CSLD3 and CESA6 expressed in Saccharomyces cerevisiae and reconstituted into proteoliposomes. Both CSLD3 and CESA6 utilized UDP-glucose but not GDP-mannose as a substrate and displayed similar substrate saturation kinetics. To confirm the nature of the polysaccharide product, radiolabeled UDP-glucose was used as a substrate. Reactions with CESA6 and CSLD3 both resulted in the time-dependent production of a radiolabeled polysaccharide that could only be digested by β-1,4-specific glucanase activity. Therefore, both enzymes produced β-1,4-glucan, the polysaccharide composing cellulose.
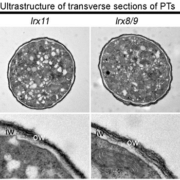
The fact that the catalytic domains are interchangeable begs the question of what is functionally different about the enzyme subunits. One possibility is that β-1,4-glucans produced by CESA proteins coalesce into microfibril bundles of 18-24 polymer strands because CESA6’s subunits organize into higher order cellulose synthase complexes. However, when the authors resolved active CESA6 and CSLD3 by size exclusion chromatography, both enzymes eluted as similarly large, approximately 700 kDa complexes. Visualization of both of these enzyme complexes with electron microscopy revealed similar 10-12nm particle with three-fold symmetry reminiscent of other previously characterized CESA complexes.
Given the similar kinetic properties and complex formation of both CESA6 and CSLD3, the authors hypothesized that a functional difference between the enzymes may arise from differing associations with cortical microtubules. Deposition of cellulose microfibrils by CESAs is often oriented according to an underlying array of microtubules. Future studies must determine whether it is the absence of cortical microtubules at the apical plasma membrane of tip growing cells, where CSLD3 activity is present, that leads to production of a β-1,4-glucan polymer that is more randomly organized than other cellulose microfibrils and better suited to tip expansion.
Brendan M O’Leary
ARC Centre of Excellence in Plant Energy Biology, University of Western Australia
ORCID: 0000-0002-8770-155X
Honkanen S, Jones VAS, Morieri G, Champion C, Hetherington AJ, Kelly S, Proust H, Saint-Marcoux D, Prescott H, Dolan L. (2016). The Mechanism Forming the Cell Surface of Tip-Growing Rooting Cells Is Conserved among Land Plants. Current Biol. 26: 3238-3244.
Park S, Szumlanski AL, Gu F, Guo F, Nielsen E. (2011). A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nature Cell Biol. 13: 973-980.
Wang X, Cnops G, Vanderhaeghen R, De Block S, Van Montagu M, Van Lijsebettens M. (2001). AtCSLD3, a cellulose synthase-like gene important for root hair growth in arabidopsis. Plant Physiol. 126: 575-586.
Yang J, Bak G, Burgin T, Barnes WJ, Mayes HB, Peña MJ, Urbanowicz B, Nielsen E. (2020). Biochemical and genetic analysis identify CSLD3 as a beta-1,4-glucan synthase that functions during plant cell wall synthesis. Plant Cell https://doi.org/10.1105/tpc.19.00637.