A new player links ABA biosynthesis and signaling with seed dormancy
Liu et al. identify new connections between known regulators of seed dormancy, acting through modulation of ABA biosynthesis and signaling. The Plant Cell (2020) https://doi.org/10.1105/tpc.20.00026
By Fei Liu and Yong Xiang, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
Background: Seed dormancy is established at the end of seed maturation and ensures that seeds sprout under favorable conditions and after they have dispersed away from the mother plant. It is a complex trait influenced by endogenous (phytohormones abscisic acid (ABA) and gibberellins) and environmental factors (including ambient temperature and relative humidity for seed hydration). A tight control of seed dormancy in crops allows uniform germination after sowing and prevents pre-harvest sprouting at the end of the growing season. However, the underlying mechanism of seed dormancy is still not fully understood.
Question: In earlier work, we had isolated the reduced dormancy5 (rdo5) mutant on the basis of its reduced seed dormancy. The RDO5 gene encode a type 2C protein phosphatase. To better understand the signaling cascade RDO5 participates in, we performed a suppressor mutant screen in the rdo5-2 background to look for additional factors controlling seed dormancy.
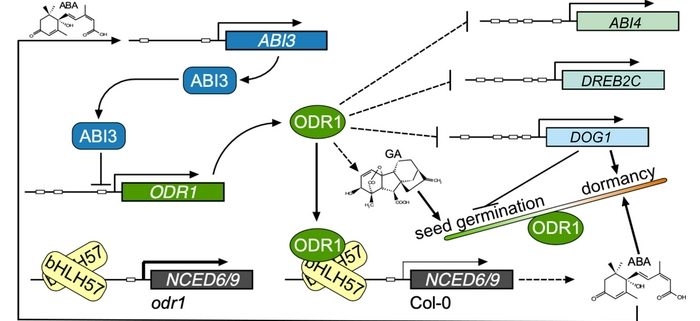
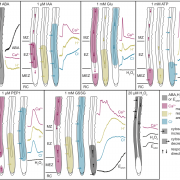
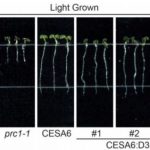
Findings: We identified two ABA-related seed dormancy regulators in Arabidopsis. One was isolated from the suppressor screen (ODR1, for reversal of rdo5, an orthologue of the rice Seed dormancy 4 Sdr4). The second protein, the basic Helix-Loop-Helix transcription factor bHLH57, interacts with ODR1. The transcription factor ABA INSENSITIVE 3 (ABI3) directly represses ODR1 transcription, thereby affecting seed dormancy by modulating ABA biosynthesis and ABA signaling. In contrast, bHLH57 imposes seed dormancy by inducing the expression of the genes encoding the ABA biosynthetic enzymes 9-CIS-EPOXYCAROTENOID DIOXYGENASE NCED6 and NCED9, and thus raises ABA levels. ODR1 interacts with bHLH57 and inhibits bHLH57-modulated NCED6 and NCED9 expression in the nucleus. Likewise, bhlh57 loss of function mutants can partially counteract the enhanced NCED6 and NCED9 expression seen in odr1 mutants and can therefore rescue their associated hyper-dormancy phenotype.
Next steps: We plan to uncover more details in the ODR1-dependent pathway, for example how ODR1 controls DOG1 expression during seed dormancy and germination.
Fei Liu et al. (2020). REVERSAL OF RDO5 1, a Homolog of rice Seed dormancy 4, Interacts with bHLH57 and Controls ABA Biosynthesis and Seed Dormancy in Arabidopsis. Plant Cell DOI: https://doi.org/10.1105/tpc.20.00026