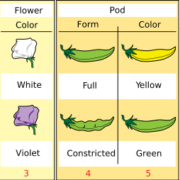
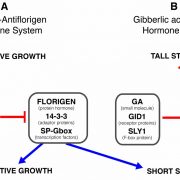
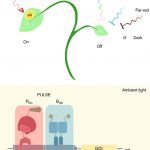
An inducible genome editing system for plants (Nature Plants)
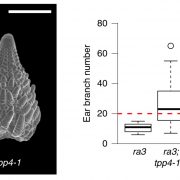
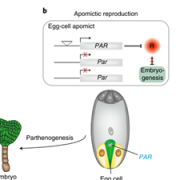
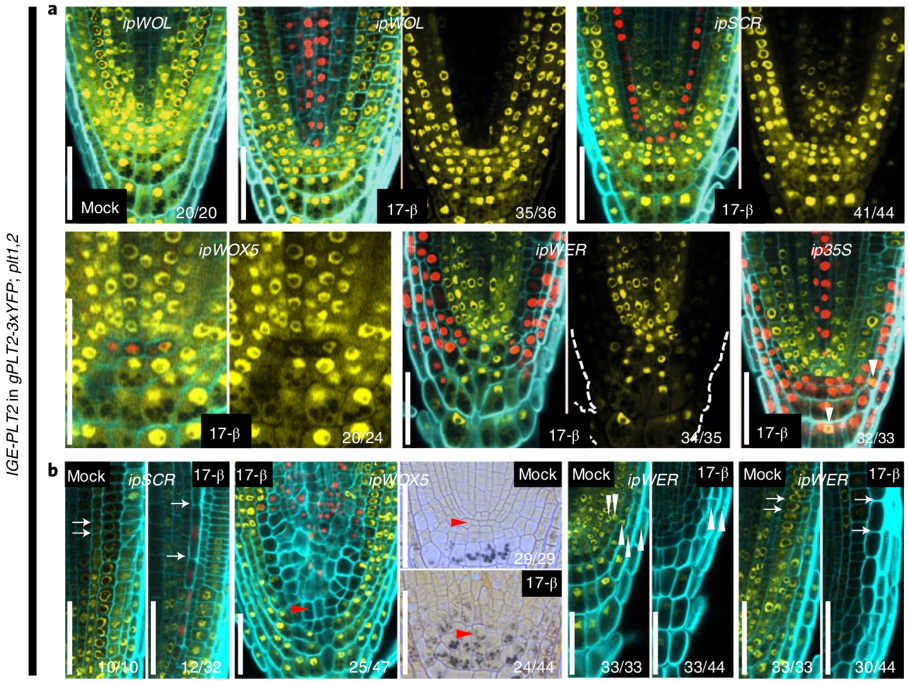 To understand gene function, it is necessary to study loss-of-function phenotypes. However, knockouts of some genes are lethal, which makes it impossible to study their loss-of-function phenotype. Although here are inducible overexpression systems and inducible knockdown systems, as yet there has been no inducible knockout system. In this paper Wang et al. developed an inducible genome editing (IGE) system by a two-step gateway cloning for conditional gene knockout. This IGE depends on the three components brought together in a plant binary vector, 1) a chimeric protein called XVE, that can be activated by estradiol and binds to the LexA binding sites in the promoter to activate the expression of the downstream gene, 2) Cas9 expressed under LexA promoter, 3) sgRNA(s) targeting the desired gene. IGE can be used in a tissue-specific or cell type-specific manner by cloning XVE under tissue-specific or cell type-specific promoters. In this paper the authors examined the effects of knocking out PLETHORA, which encodes an AP2/EREBP transcription factor, RETINOBLASTOMA-RELATED (RBR), which encodes a cell cycle regulator, and GNOM, which encode a brefeldin A sensitive ARF guanine-nucleotide exchange factor. The authors confirmed their conditional knockouts in both a broad domain and cell type-specific domain, and they compared IGE with inducible artificial microRNAs and showed that IGE is more specific. In a nutshell, this paper provides a system for efficient, precise inducible knockout of genes in Arabidopsis. (Summary by Vijaya Batthula @Vijaya_Batthula) Nature Plants 10.1038/s41477-020-0695-2
To understand gene function, it is necessary to study loss-of-function phenotypes. However, knockouts of some genes are lethal, which makes it impossible to study their loss-of-function phenotype. Although here are inducible overexpression systems and inducible knockdown systems, as yet there has been no inducible knockout system. In this paper Wang et al. developed an inducible genome editing (IGE) system by a two-step gateway cloning for conditional gene knockout. This IGE depends on the three components brought together in a plant binary vector, 1) a chimeric protein called XVE, that can be activated by estradiol and binds to the LexA binding sites in the promoter to activate the expression of the downstream gene, 2) Cas9 expressed under LexA promoter, 3) sgRNA(s) targeting the desired gene. IGE can be used in a tissue-specific or cell type-specific manner by cloning XVE under tissue-specific or cell type-specific promoters. In this paper the authors examined the effects of knocking out PLETHORA, which encodes an AP2/EREBP transcription factor, RETINOBLASTOMA-RELATED (RBR), which encodes a cell cycle regulator, and GNOM, which encode a brefeldin A sensitive ARF guanine-nucleotide exchange factor. The authors confirmed their conditional knockouts in both a broad domain and cell type-specific domain, and they compared IGE with inducible artificial microRNAs and showed that IGE is more specific. In a nutshell, this paper provides a system for efficient, precise inducible knockout of genes in Arabidopsis. (Summary by Vijaya Batthula @Vijaya_Batthula) Nature Plants 10.1038/s41477-020-0695-2