Alternative Splice Variant Sheds Light on Temperature Acclimation in Algae
Climate change is a potent environmental force that all living organisms must contend with. This is especially true for photosynthetic, microalgae that are forced to acclimate to ever changing water temperatures and coordinate changes in their physiology and growth rates (Singh and Singh, 2015). These adjustments can have profound impacts on carbon fixation on Earth and the acceleration of climate change (Schaum et al., 2017).
Microalgae, such as the model species Chladydomonas reinhardtii (C. reinhardtii) also provide a useful system to study temperature sensing and signaling by having well-defined growth conditions that can be carefully controlled (Merchant et al., 2007). C. reinhardtii is known as a “cosmopolitan species” capable of growing in physiological temperatures ranging from 18C (lower temp) to 28C (higher temp) and optimally at 25C (ambient temp). Although not initially lethal, growth at lower and higher temps requires acclimation for physiological adjustment and survival, even if brought back to ambient temperatures (Vítová et al., 2011).
C. reinhardtii uses an RNA-binding protein, called CHLAMY1 to integrate temperature acclimation within physiological temperatures to the circadian clock (clock) (Voytsekh et al., 2008). CHLAMY1 does this through its two subunits, C1 and C3. C1 contains K homology (KH) RNA-binding domains that become hyperphosphorylated under lower temperatures, while C3 contains RNA recognition motifs (RRMs) and its protein is upregulated at lower temperatures. XRN1 (or ROC86), a known clock component interacts with C3, and is also upregulated under lower temps (Dathe et al., 2012). The interaction of C3 and XRN1 provides a direct integration of temperature and clock regulation, but may also involve other regulatory factors.
Li et al. (this issue) sought to identify other factors working with CHLAMY1 by conducting a co-immunoprecipitation (Co-IP) experiment using C. reinhardtii extract and XRN1 as bait. In addition to C3, the authors discovered a number of novel interacting RNA-binding proteins, including one called Musashi. Musashi proteins are known to fulfill a number of roles in Drosophila and other animal species, such as translational control, polyadenylation and signaling but are poorly understood in non-animal systems (Weill et al., 2017). The Musashi gene model (Cre16.g662702.t1.1) predicts a 77 kDa protein (77 kDa) with two RRMs and two transmembrane domains (TMDs). This gene model was used to develop an antibody and probe C. reinhardtii extract. A number of Musashi-like protein species were identified and ranged in size from 57 to 78 kDa, suggesting degradation products or splice variants.
To investigate the source of the additional Musashi-like protein species, the authors used the 77 kDa sequence to mine available transcriptome data, revealing 25 splice variants (variants). The 25 novel variants spanned the 77 kDa coding sequence and contained up to three RRMs and two TMDs with differences in both N and C-termini. Only one variant (60 kDa) was highly expressed and contained all three RRM and TMD domains. A pull-down assay was used to test the interaction of the 25 variants and XRN1. The highest number of variants identified contained all three RRMs, although the 77 kDa variant containing only two RRMs also interacted with XRN1 suggesting the importance of the N-terminal region for protein-protein interactions.
In addition to being capable of binding XRN1, the 60 kDa variant was also able to bind C3 and (UG)7 repeats within a mRNA 3’UTR, a function known to Musashi proteins and C3 itself. The RNA binding properties of Musashi and C3 are important to function of the complex since XRN1 does not bind (UG)7 repeats, and XRN1 is hypothesized to act downstream through subcellular localization of target transcripts and regulating expression (Dathe et al., 2012). Furthermore, the 60 kDa variant contains three RRMs and bound its target RNA more tightly compared to variants with less RRMs, regardless of size. This is in contrast with animal Musashi proteins which use only two RRMs and suggests a divergence in function from the animal Musashi proteins (Zhao et al., 2004).
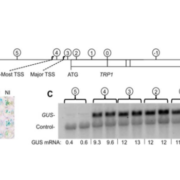
CHLAMY1 integrates temperature information through differential expression and post-translational processing of its two subunits, C1 and C3, and its binding affinity to (UG)7 repeats, with a lower affinity near midday (LD4) and a higher affinity at early night (LD14) (Voytsekh et al., 2008). Direct interaction between C3 and the 60 kDa Musashi variant suggested the 60 kDa variant may also be regulated by temperature. This was tested through an expressional analysis at the protein and transcript level. As expected, C3 protein was up-regulated at the lower temp, especially near midday. On the other hand, XRN1 and the 60 kDa variant protein were up-regulated at the higher temp, especially during early night (Figure 1). Interestingly, the 60 kDa variant transcript was largely the same between the two time periods, suggesting clock regulation at the post-transcriptional level.
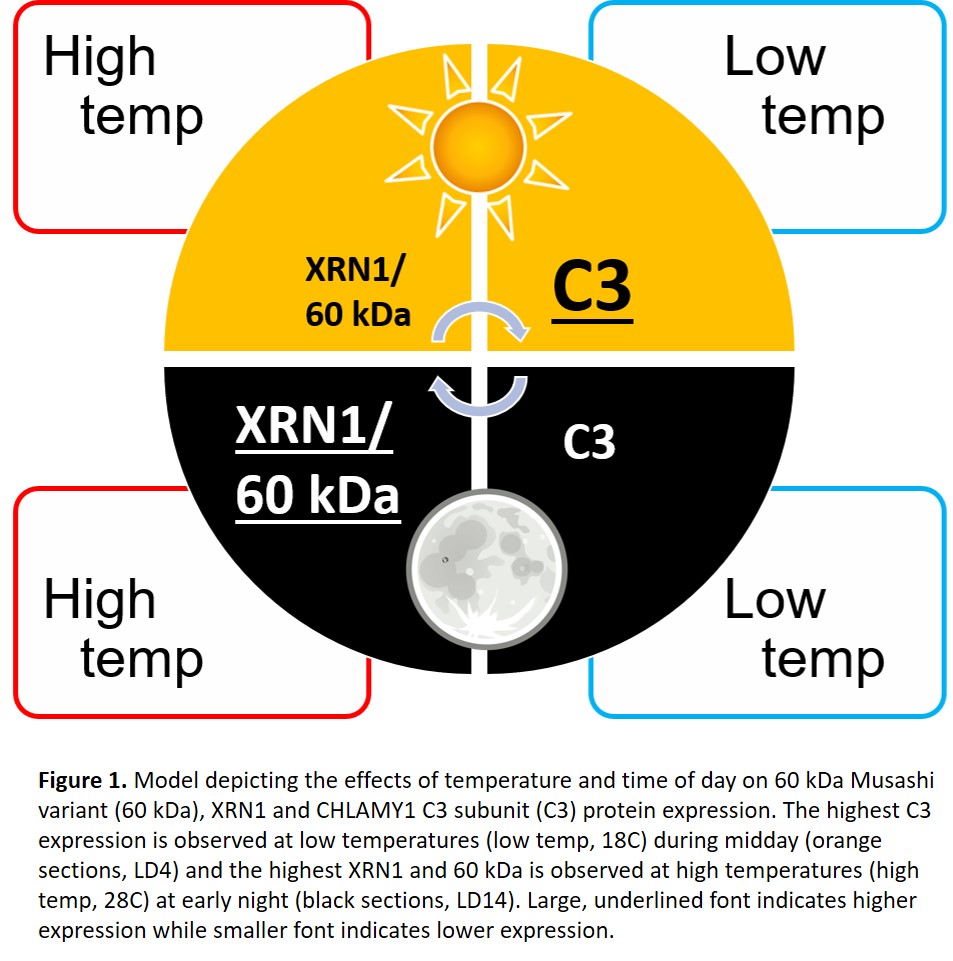
Temperature-specific regulation of XRN1, C3 and the 60 kDa Musashi variant could mean each component is active during temperature acclimation and survival following exposure to higher and lower temps. This function was tested using a thermo-survival assay which exposes cultures to 12-hours of dark at the lower and higher temps as well as heat shock (42C) and near heat shock (37C) conditions. Knock-out and artificial microRNA (amiRNA) knock-down mutants of XRN1 showed significantly reduced survival at all temperatures tested except 42C, where most cells were killed regardless of treatment. This relationship was also observed with RNA interference (RNAi) knock-down mutants of C3, suggesting both XRN1 and C3 are critical to temperature acclimation, especially during temperature stress. Conversely, overexpression of the 60 kDa Musashi variant resulted in reduced survival only within physiological temperatures, with no significant differences under near heat shock conditions.
Understanding mechanisms of temperature acclimation is becoming more important with changes in global climate and increases in ocean temperatures. Li et al. have identified a new component in the temperature acclimation system in microalgae through characterization of an alternatively spliced variant, Musahshi 60 kDa and have demonstrated its function under physiological conditions. The putative roles of the other Musashi alternatively spliced variants are unknown but common to RNA-binding proteins (Cho et al., 2015). Further investigation is warranted to interrogate their function as well as downstream RNA targets.
LITERATURE CITED
Cho SK, Sharma P, Butler NM, Kang I-H, Shah S, Rao AG, Hannapel DJ (2015) Polypyrimidine tract-binding proteins of potato mediate tuberization through an interaction StBEL5 RNA. J. Expt. Botany 66: 6835-6847
Dathe H, Prager K, Mittag M (2012) Novel interaction of two clock-relevant RNA-binding proteins C3 and XRN1 in Chlamydomonas reinhardtii. FEBS Lett 586: 3969-3973
Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245-250
Schaum C-E, Barton S, Bestion E, Buckling A, Garcia-Carreras B, Lopez P, Lowe C, Pawar S, Smirnoff N, Trimmer M, et al (2017) Adaptation of phytoplankton to a decade of experimental warming linked to increased photosynthesis. Nat Ecol Evol 1: 94
Singh SP, Singh P (2015) Effect of temperature and light on the growth of algae species: A review. Renewable Sustainable Energy Rev 50: 431-444
Vitova M, Bisova K, Umysova D, Hlavova M, Kawano S, Zachleder V, et al. (2011) Chlamydomonas reinhardtii: duration of its cell cycle and phases at growth rates affected by light intensity. Planta 233: 75–86
Voytsekh O, Seitz SB, Iliev D, Mittag M (2008) Both subunits of the circadian RNA-binding protein CHLAMY1 can integrate temperature information. Plant Physiol 147: 2179–2193
Weill L, Belloc E, Castellazzi CL, Méndez R (2017) Musashi 1 regulates the timing and extent of meiotic mRNA translational activation by promoting the use of specific CPEs. Nat Struct Mol Biol 24: 672-681
Zhao B, Schneid C, Iliev D, Schmidt E-M, Wagner V, Wollnik F, Mittag M (2004) The circadian RNA-binding protein CHLAMY 1 represents a novel type heteromer of RNA recognition motif and lysine homology domain-containing subunits. Eukaryot Cell 3: 815-825