Accurate and versatile 3D segmentation of plant tissues at cellular resolution (bioRxiv)
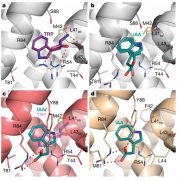
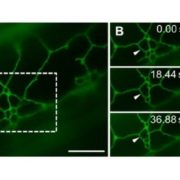
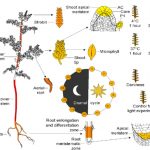
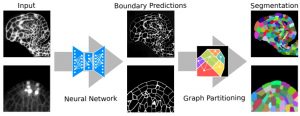 Advances in microscopy allow biologists to document and understand phenotypic changes at the tissue level. Despite this advancement, analysis of microscopic images to characterize changes in phenotypes is still under development. In this paper, Wolny et al. have developed an image analysis tool PlantSeg that enables users to analyze microscopic images from confocal and light-sheet microscopy to perform segmentation on the tissue of interest. Segmentation in images of multicellular organisms like plants requires high-resolution images with low background signal and clear boundaries of the cells, which requires imaging expertise. In this paper, the authors have developed a tool that can predict clear boundaries using a convolution neural network followed by segmentation for the analysis of cell shape, volume, and patterning. To evaluate the tools, the authors successfully performed segmentation using fixed tissue of Arabidopsis ovules and also time-lapse development of lateral root primordia in Arabidopsis. This free open-source tool is available for users and further information can be found in https://github.com/hci-unihd/plant-seg. In addition to the GUI, for advanced users with knowledge of Python the source code is made available in the same link. (Summary by Suresh Damodaran) bioRxiv 10.1101/2020.01.17.910562
Advances in microscopy allow biologists to document and understand phenotypic changes at the tissue level. Despite this advancement, analysis of microscopic images to characterize changes in phenotypes is still under development. In this paper, Wolny et al. have developed an image analysis tool PlantSeg that enables users to analyze microscopic images from confocal and light-sheet microscopy to perform segmentation on the tissue of interest. Segmentation in images of multicellular organisms like plants requires high-resolution images with low background signal and clear boundaries of the cells, which requires imaging expertise. In this paper, the authors have developed a tool that can predict clear boundaries using a convolution neural network followed by segmentation for the analysis of cell shape, volume, and patterning. To evaluate the tools, the authors successfully performed segmentation using fixed tissue of Arabidopsis ovules and also time-lapse development of lateral root primordia in Arabidopsis. This free open-source tool is available for users and further information can be found in https://github.com/hci-unihd/plant-seg. In addition to the GUI, for advanced users with knowledge of Python the source code is made available in the same link. (Summary by Suresh Damodaran) bioRxiv 10.1101/2020.01.17.910562