A Kinase- and Proteasome-Mediated Link Between Lipid Biosynthesis and Energy Homeostasis
IN BRIEF by Nancy R. Hofmann nhofmann@aspb.org
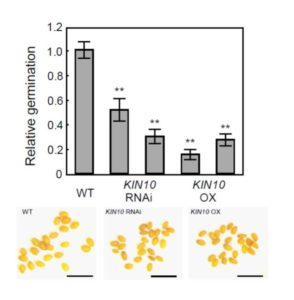 The economy of living cells includes energy production, energy utilization, and energy storage. Thus, a plant’s energy budget must connect the decision to produce lipids (for energy storage) with its overall energy status. Energy sensor kinases, such as KIN10 and KIN11 in Arabidopsis thaliana, are key transcriptional regulators that integrate energy and stress signaling networks (reviewed in Sheen, 2014). New work from Zhai et al. (2017) shows that they also connect energy status with the biosynthesis of lipid storage molecules.
The economy of living cells includes energy production, energy utilization, and energy storage. Thus, a plant’s energy budget must connect the decision to produce lipids (for energy storage) with its overall energy status. Energy sensor kinases, such as KIN10 and KIN11 in Arabidopsis thaliana, are key transcriptional regulators that integrate energy and stress signaling networks (reviewed in Sheen, 2014). New work from Zhai et al. (2017) shows that they also connect energy status with the biosynthesis of lipid storage molecules.
Given previous reports of seemingly conflicting effects of KIN10 overexpression on seeds— enhancing seed development and oil contents, but depressing seed germination rates— Zhai et al. revisited the question of what roles KIN10 plays in seeds. The authors compared seeds of KIN10 overexpression lines to those of wild type and KIN10-suppressed (RNAi) lines. Both transgenic lines, whether up- or down-regulated for KIN10, had smaller seeds than wild type, and both had poor germination compared to WT (see Figure 1), with significantly decreased levels of the fatty acid triacylglycerol (TAG).
This effect on TAG levels prompted Zhai et al. to probe the effects of KIN10 on WRINKLED1 (WRI1), a conserved regulator of TAG accumulation (Grimberg, 2015). Co-expression of Arabidopsis WRI1 and OLEOSIN1 (OLE1) in N. benthamiana leaves induces TAG accumulation by increasing fatty acid biosynthesis and storage lipid packaging. Zhai et al. found that this lipid accumulation was suppressed when WRI1 and OLE1 were co-expressed with KIN10 (or its close homolog KIN11). In the presence of KIN10, less WRI1 accumulated in N. benthamiana leaves, a decrease that was dependent on the proteasomal pathway and on an active kinase domain in KIN10. Zhai et al. found direct interaction of KIN10 (or KIN11) with WRI1 in the nucleus. Further, in vitro kinase assays showed that KIN10, in the presence of its activator GRIK1, could phosphorylate WRI1.
The authors went on to show that KIN10 likely phosphorylates WRI1 at two sites, each in a variant of the canonical KIN10 phosphorylation motif. Intriguingly, each of the WRI1 phosphorylation sites overlapped with an AP2 DNA binding domain, and when WRI1 target DNA was present in the kinase assay, phosphorylation of WRI1 was decreased. Phosphorylated WRI1 was degraded by Arabidopsis cell extracts more quickly than the non-phosphorylated form.
The results from Zhai et al. support a model in which WRI1, as a master regulator of TAG biosynthesis, is targeted for degradation by the proteasome pathway upon phosphorylation by the energy sensor KIN10. As KIN10 activity is negatively regulated by glucose, TAG biosynthesis would be favored when glucose is abundant. Thus, this work reveals an important mechanistic connection in energy homeostasis.
REFERENCES
Grimberg, A., Carlsson, A.S., Marttila, S., Bhalerao, R., and Hofvander, P. (2015). Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol 15, 192.
Sheen, J. (2014). Master Regulators in Plant Glucose Signaling Networks. Journal of Plant Biology 57, 67-79.
Zhai, Z., Liu, H., and Shanklin, J. (2017). Phosphorylation of WRINKLED1 by KIN10 Results in its Proteasomal Degradation, Providing a Link Between Energy Homeostasis and Lipid Biosynthesis. Plant Cell. doi: 10.1105/tpc.17.00019.




Leave a Reply
Want to join the discussion?Feel free to contribute!