Questions about Coenzyme Q? A New Genetic/Metabolic Study Has Answers
Anyone who works on plants should be dazzled by the complexity and versatility of plant metabolism. In fact, why restrict this to plant biologists? We all love plant metabolites, whether we’re enjoying the caffeine in our morning cup of tea, or the capsaicin heat in the pepper flakes on our lunchtime pizza. One metabolite we want to show a little love for is ubiquinone, also known as Coenzyme Q. Ubiquinone is a benzoic acid compound (reviewed in Wildham et al., 2015) that functions in electron transport at the inner mitochondrial membrane. In addition to its function in respiration, ubiquinone acts as an antioxidant to protect membrane lipids from damage by free radicals, which are often produced during respiration.
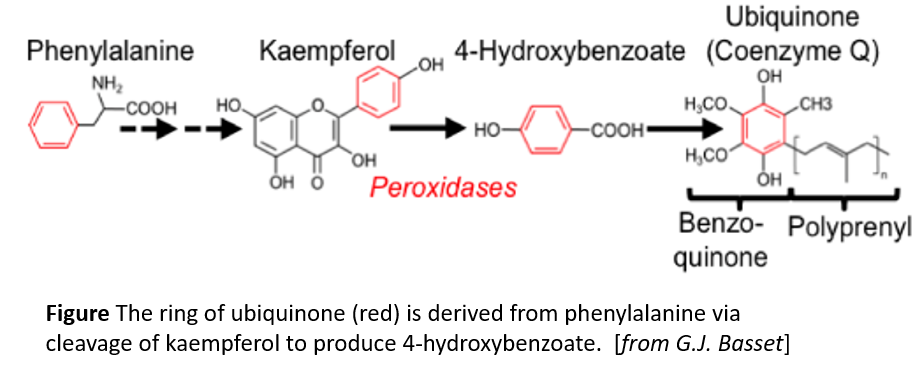
Ubiquinone contains a benzoquinone ring and a polyprenyl tail (see figure). Moreover, (reviewed in Liu and Lu, 2016) it has a short half-life, on the order of a day or so, so plants need to make new ubiquinone continuously. In contrast to animals and fungi, plants can get the benzoquinone ring of ubiquinone from phenylalanine (Block et al., 2014). However, the pathway and responsible enzymes had long remained unclear. Now, Soubeyrand et al. (2018) bring some serious genetic and metabolic analysis to bear on this question. The authors first used metabolic labeling experiments, feeding phenylalanine-[Ring–13C6] to various Arabidopsis thaliana mutants that are affected in different steps of the biosynthesis of the precursor 4-hydroxybenzoate. The authors constructed a gene interaction network and hierarchical clustering analysis to identify additional players in this pathway, starting with known ubiquinone biosynthetic genes. These analyses revealed a link to flavonoid metabolism in the production of ubiquinone from phenylalanine. Quantification of ubiquinone levels in Arabidopsis flavonoid biosynthetic pathway mutants confirmed the importance of the flavonoid pathway and showed that the pathways diverged after the reaction catalyzed by flavanone-3-hydroxylase (F3H). In agreement with this, a tomato (Solanum lycopersicum) mutant affecting F3H had reduced ubiquinone contents compared with wild type.
The authors next turned their attention to finding key metabolic intermediates. The identification of F3H as a key enzyme eliminated downstream flavonoid intermediates such as anthocyanins, but put the spotlight on two related metabolites, dihydrokaempferol and kaempferol. Feeding experiments with the Arabidopsis f3h mutant and 13C6-labeled precursors showed that ubiquinone gets its benzoquinone ring from kaempferol (figure). Moreover, the authors hypothesized that, since plant tissues tend to have peroxidases that affect flavonoids, a peroxidase might be responsible for removing the benzene ring from kaempferol. Indeed, reactions with Arabidopsis protein extracts showed H2O2-dependent release of 4-hydroxybenzoate from kaempferol and inactivation of heme-containing enzymes by azide prevented this reaction. Therefore, peroxidases likely release 4-hydroxybenzoate from kaempferol; this ring is then incorporated into ubiquinone. This answers a longstanding question about ubiquinone biosynthesis.
If this novel metabolic twist of making an essential cofactor, ubiquinone, from kaempferol, which had been considered a secondary metabolite, were not cool enough, these findings have implications for plant stress tolerance, and even human nutrition. As a lipid-soluble antioxidant, ubiquinone can help plants prevent damage from reactive oxygen species produced under abiotic stress conditions. Metabolic engineering of ubiquinone to produce plant varieties with improved stress tolerance has raised substantial interest and ubiquinone’s antioxidant properties have promoted its use in human dietary supplements, and even in anti-aging skin creams. Therefore, metabolic engineering to produce plants with increased ubiquinone has potential importance for agriculture and medicine. Indeed, the authors find that increasing kaempferol biosynthesis increased the ubiquinone contents of tomato fruits. Moreover, they suggest that identifying and knocking out the enzymes that add sugars to kaempferol and thus prevent its cleavage will further enhance ubiquinone biosynthesis. So, let’s raise a glass or cup of our favorite plant products to this versatile and interesting plant cofactor.
REFERENCES
Block, A., Widhalm, J.R, Fatihi,A., Cahoon, R.E., Wamboldt, Y., Elowsky, C., Mackenzie, S.A., Cahoon, E.B, Chapple, C., Dudareva, N., and Basset, G.J (2014) The origin and biosynthesis of the benzenoid moiety of ubiquinone (Coenzyme Q) in Arabidopsis. Plant Cell 26: 1938-1948.
Liu, M., and Lu, S. (2016) Plastoquinone and Ubiquinone in Plants: Biosynthesis, Physiological Function and Metabolic Engineering. Front Plant Sci. 2016; 7: 1898. DOI: 10.3389/fpls.2016.01898
Soubeyrand, E., Johnson, T.S., Latimer, S., Block, A., Kim, J., Colquhoun, T.A., Butelli, E., Martin, C., Wilson, M.A., and Basset, G.J. (2018) The Peroxidative Cleavage of Kaempferol Contributes to the Biosynthesis of the Benzenoid Moiety of Ubiquinone in Plants. Plant Cell DOI: https://doi.org/10.1105/tpc.18.00688.
Widhalm, J.R., and Dudareva, N. (2015). A familiar ring to it: biosynthesis of plant benzoic acids. Mol. Plant. 8: 83-97.