How resurrection plants survive being hung out to dry
Resurrection plants have the unique ability to survive extreme dehydration (desiccation), lying dormant for months or sometimes years until rehydration is possible. This formidable survival strategy has independently evolved several times across the land plant phylogeny, and several phylogenetically diverse resurrection plant genomes have been sequenced and assembled in an attempt to understand the causal genetic mechanisms. Large-scale comparisons across each of these phylogenetically distant resurrection plant genomes reveals that some conserved molecular signatures may underlie desiccation tolerance (Illing et al., 2005; Zhang and Bartels, 2018), but overall the genes, networks, and regulatory factors that underlie desiccation tolerance remain largely unknown.
Here, VanBuren et al. take a powerful phylogenetic comparative approach to uncover the genetic basis for desiccation tolerance in Lindernia, a eudicot in the Lamiales order that contains a range of desiccation tolerance diversity. They approach the question by generating genome assemblies for two closely related species of Lindernia – one that is desiccation tolerant (Lindernia brevidens) and another that is desiccation sensitive (Lindernia subracemosa). By combining long PacBio reads for both genomes with in vivo chromatin conformation capture (Hi-C) for L. brevidens, both genome assemblies were highly contiguous and ordered and oriented onto 14 chromosomes each.
Next, the authors used a series of comparative genomic analyses to uncover the evolutionary history of these genomes. Both genomes are largely collinear with each other, and analysis of the syntenic blocks of shared collinear genes (syntelogs) revealed an ancient polyploidy event, forming a hypothesis in the manuscript that these two genomes have retained or lost different sets of paralogous genes following whole-genome duplication that may underlie desiccation tolerance and other lineage-specific traits. To identify these possible patterns of gene duplication and loss following the whole-genome duplication event, the authors used the genome collinearity information to determine whether a gene was retained in duplicate or lost in one genome. Indeed, the lineage-specific genes that were retained in desiccation-tolerant L. brevidens are enriched for some suggestive gene ontology (GO) terms like heat response and chlorophyll biosynthesis that may be involved in desiccation tolerance.
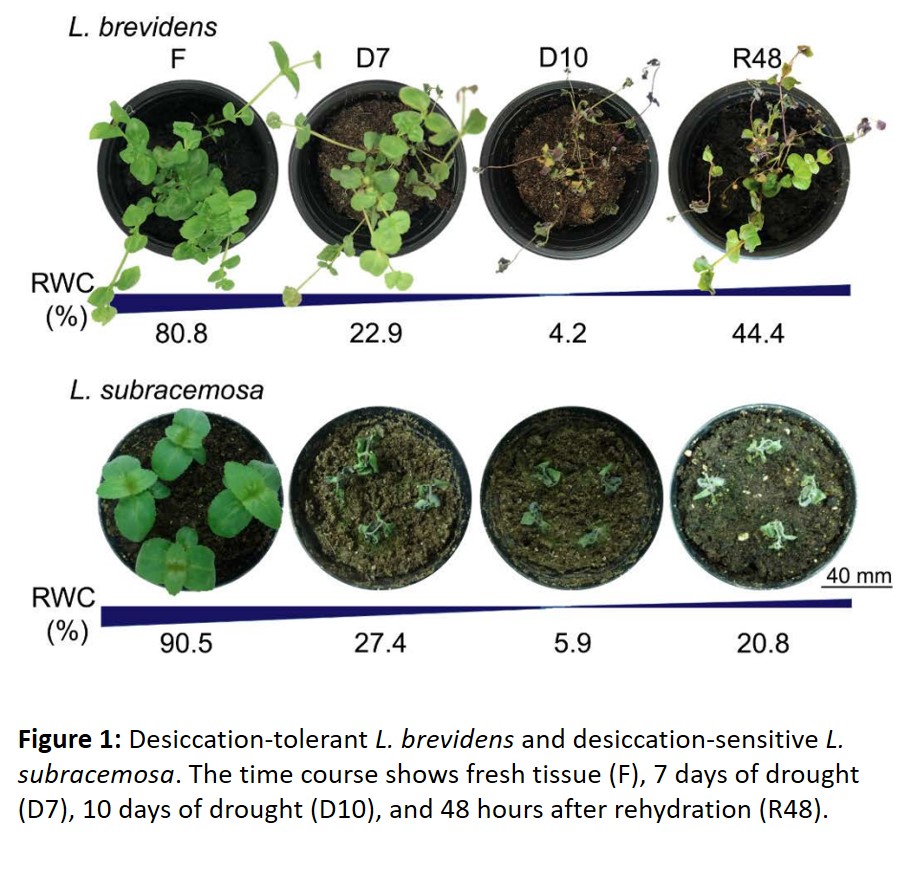
While comparative genomics of the whole genomes revealed some clues about the differentiation between L. brevidens and L. subracemosa, the authors then leveraged a powerful RNA-seq experiment over a time-course of severe desiccation and rehydration. As expected, after desiccation and rehydration the L. subracemosa plants were dead, yet the L. brevidens plants were mostly viable and active within 48 hours after watering (Figure 1). In some particularly stressful stages, such as from severe dehydration to total desiccation, there were very few differentially expressed genes in L. brevidens, and quite the opposite in the L. subracemosa, suggesting that the desiccation-tolerant L. brevidens displays transcriptional stability during water stress. To identify gene modules or networks that specifically underlie desiccation tolerance, the authors took a gene correlation network approach which revealed perhaps the most powerful insight of the manuscript: that during dehydration and desiccation, both species expressed a very different complement of co-expressed genes, meaning that there is a fundamental gene regulatory network rewiring that occurred between these two species. In particular, the desiccation-tolerant L. brevidens upregulated a cluster of genes enriched in ABA-responsive and seed-specific elements that are already associated with dehydration.
This combination of genomics, physiology, and elegant experimental design brings to light how polyploidy and subsequent diploidization can rewire entire stress response pathways. VanBuren et al. reinforce that understanding the molecular basis of complex phenotypes requires a creative experimental and analytical approach. Their comparative phylogenetic approach in Lindernia has revealed a potentially novel mechanism for desiccation tolerance, and future work will continue to focus on the regulatory mechanisms that underlie this peculiar stress adaptation.
REFERENCES
Illing, N., Denby, K.J., Collett, H., Shen, A., and Farrant, J.M. (2005). The signature of seeds in resurrection plants: a molecular and physiological comparison of desiccation tolerance in seeds and vegetative tissues. Integr. Comp. Biol. 45: 771–787.
VanBuren, R., Wai, C.M., Pardo, J., Giarola, V., Ambrosini, S., Song, X., and Bartels, D. (2018) Genomics of desiccation tolerance in Lindernia. Plant Cell 10.1105/tpc.18.00517
Zhang, Q. and Bartels, D. (2018). Molecular responses to dehydration and desiccation in desiccation-tolerant angiosperm plants. J. Exp. Bot. 69: 3211–3222.



