A community repository of plant illustrations
Guest post by Erin Sparks, Guillaume Lobet, Larry York and Frédéric Bouché
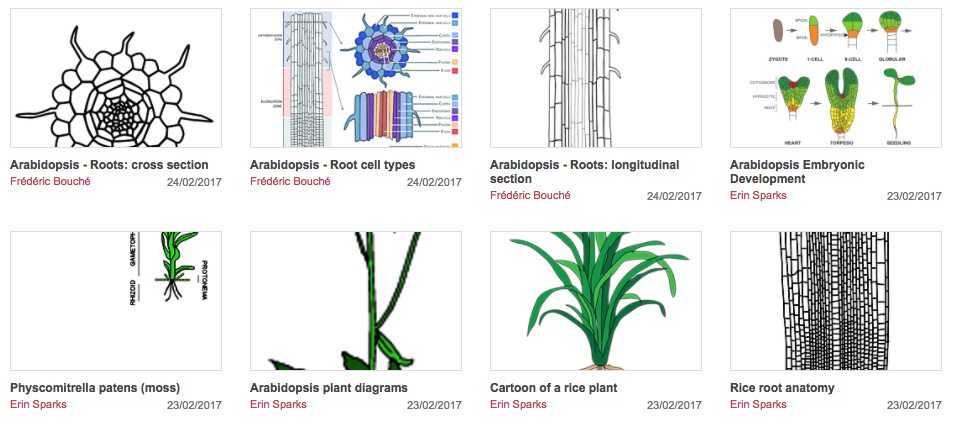
It is midnight on a cold winter evening and you are scheduled to give a seminar at 8 am the next morning. All you are missing to complete your presentation is one last graphic to illustrate your conclusions. You wearily open Adobe Illustrator, stare at a blank artboard and think “isn’t there a better way?” What if there was a place where you could access community plant illustrations to use or modify? Good news! A Plant Illustrations Repository is now available to the plant scientific community!
In a time when scientific communication is becoming increasingly important, one method of communication is through illustrations and graphics. For example, graphical abstracts are now frequently employed by journals to summarize the contents of a manuscript and provide a visual overview of the work. These graphics are generated in programs such as Adobe Illustrator, Inkscape, or even PowerPoint. For additional information about graphical abstracts, we refer the reader to this blog post and a recent webinar by Fred entitled: “Communicate your Research Efficiently Using Graphical Abstracts” hosted by Plantae and the American Society of Plant Biologists.
In an effort to promote scientific communication and the use of graphical abstracts, we have initiated a repository for community-generated pictures and vector graphic illustrations of plants. It takes a lot of effort to make vector graphics so we might as well share them among the community. Thus our goal was to provide a central location where plant scientists can contribute their images and graphics to be used or modified by others (and you get credit for it!).
We decided to use a figshare Collection for a couple of reasons:
- It is free to use and any scientist can deposit data there in a couple of clicks.
- One dataset can contain more than one file (for instance, .ai + .png + .pdf).
- It provides unlimited storage for data that are made public.
- It provides each public dataset with a doi (digital online identifier), which make citation possible, even for a single figure.
- Finally, the Collection feature of figshare allows us to collect images and illustrations from multiple accounts (yours!) into a single place and to present it nicely!
We invite you to visit the Plant Illustrations Repository to learn more about contributing and utilizing this resource.
Happy Communicating!
Erin, Guillaume, Larry and Fred
Erin Sparks is a Postdoc with Philip Benfey at Duke University and her recent work focuses on understanding the development and function of maize brace roots. You can follow Erin on Twitter @ErinSparksPhD
Guillaume Lobet is an Associate Professor at the Forschungszentrum Jülich in Germany and the Université Catholique de Louvain in Belgium. His work focuses on the development of whole plant models. You can follow Guillaume on Twitter @guillaumelobet
Larry York is an Assistant Professor at The Samuel Roberts Noble Foundation in Ardmore, OK, USA. His work focuses on root functional phenomics including high-throughput phenotyping of root system architecture and physiology. You can follow Larry on Twitter @LarryMattYork
Frédéric Bouché is a Postdoc with Rick Amasino at the University of Wisconsin-Madison. His work focuses on the identification of the genetic mechanisms controlling the timing of flowering in the model grass Brachypodium, a species closely related to wheat, barley, and oats. You can follow Fred on Twitter @Frederic_Bouche
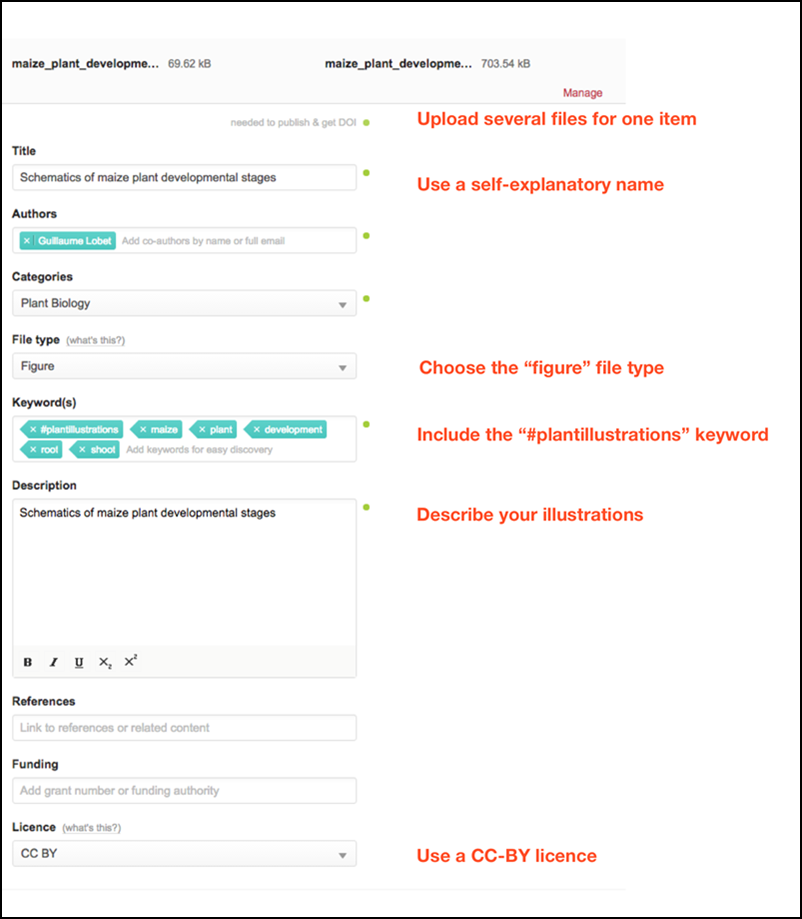
How to submit:
- Log in or sign up to your figshare account (https://figshare.com/ )
- Upload your illustration (details here)
- Upload as many files as you want for the same item (e.g. different file types of the same image)
- Fill out the title and description
- Use the “Figure” file type
- Add the “#plantillustrations” tag (with the “#”)
- Publish your figure
- Send us an email, so we can add your work to the collections



Leave a Reply
Want to join the discussion?Feel free to contribute!