A cis-regulatory atlas in maize at single-cell resolution (bioRxiv)
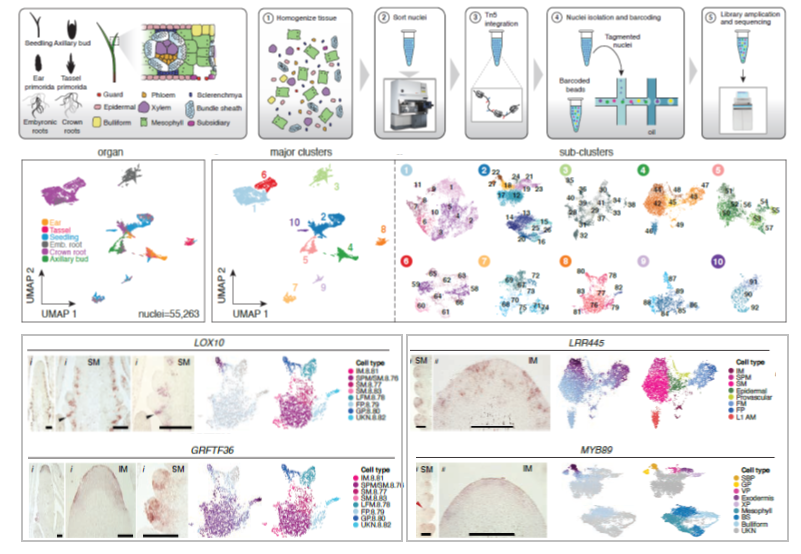
Cis-regulatory elements (CREs) are DNA sequences found near or within genic regions that drive proper gene expression in time and space. Thus, CREs play essential roles in the diversification of spatially distinct cell-types with specialized function in multicellular organisms. To identify CREs underlying the specification of different organs in the crop species Zea mays (maize), Marand and colleagues improved a protocol for single-cell sequencing of Assay for Transposase Accessible Chromatin (scATAC-seq). By optimizing a method to group nuclei with similar patterns of chromatin accessibility, they identified 10 clusters composed of nuclei from the same organ and 92 sub-clusters associated with 165,913 Accessible Chromatin Regions (ACRs) and 52 known cell-types. Validation of the mRNA spatio-temporal distribution of selected genes by in situ hybridization supports the use of ACRs to predict gene expression. The authors found an association between cluster-specific ACRs and enhancer activity. Interestingly, almost half of the ACRs located in distal regions, and a great proportion overlapped with transposable elements – including the enhancer of the domestication gene teosinte branched1. These findings reinforce the functional importance of transposons in the evolution of CREs within accessible chromatin, which has contributed to phenotypic variation in maize. The authors also demonstrated that the selection of agronomic traits related to inflorescence architecture has targeted polymorphisms of ACRs specific for floral cells. Furthermore, they used ACRs and transcription factors binding sites to predict cell-type identity and function based on the dynamics of chromatin accessibility. The authors envision this comprehensive framework as valuable tool for crop genetic improvement through genome editing and synthetic biology. (Summary and image adaptation by Michela Osnato @michela_osnato) bioRxiv 10.1101/2020.09.27.315499