Widespread long-range cis-regulatory elements in the maize genome ($) (Nature Plants)
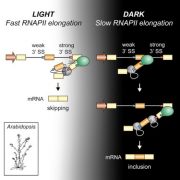
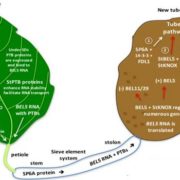
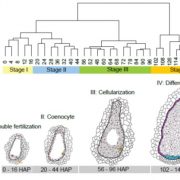
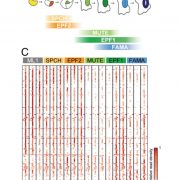
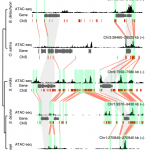
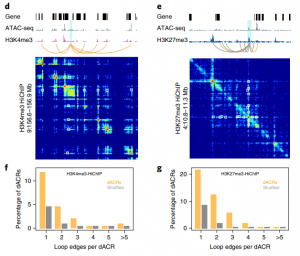 The chromatin landscape is key in the regulation of gene expression. Many studies have been made in Arabidopsis, but its genomic architecture is different to other plants. Here Ricci et al. present a very comprehensive study of chromatin analysis in maize. This work includes a huge amount of sequencing data, including ATAC-seq (transposase-accessible chromatin sequencing) chromatin accessibility landscape, DAP-seq for 32 maize transcription factor genome-wide binding sites, 9 ChiP-seq for histone features, and loop analysis by HiC and HiChiP. In maize, one-third of the accessible chromatin regions are located far from gene transcription starting sites (>2kb). These regions are enriched in transcription factor binding sites. Moreover, different correlation analysis suggests they are functional and important for gene regulation. In some cases, these distal regions form chromatin loops mediated by transcription factors and histone modifications. Putative enhancer activity assessed in protoplasts also supports the hypothesis that they are functional. Together with a companion paper by Lu et al. (see here), these works are opening doors to study distal enhancers in plants and for comparison to other eukaryotes. (Summary by Facundo Romani) Nature Plants 10.1038/s41477-019-0547-0
The chromatin landscape is key in the regulation of gene expression. Many studies have been made in Arabidopsis, but its genomic architecture is different to other plants. Here Ricci et al. present a very comprehensive study of chromatin analysis in maize. This work includes a huge amount of sequencing data, including ATAC-seq (transposase-accessible chromatin sequencing) chromatin accessibility landscape, DAP-seq for 32 maize transcription factor genome-wide binding sites, 9 ChiP-seq for histone features, and loop analysis by HiC and HiChiP. In maize, one-third of the accessible chromatin regions are located far from gene transcription starting sites (>2kb). These regions are enriched in transcription factor binding sites. Moreover, different correlation analysis suggests they are functional and important for gene regulation. In some cases, these distal regions form chromatin loops mediated by transcription factors and histone modifications. Putative enhancer activity assessed in protoplasts also supports the hypothesis that they are functional. Together with a companion paper by Lu et al. (see here), these works are opening doors to study distal enhancers in plants and for comparison to other eukaryotes. (Summary by Facundo Romani) Nature Plants 10.1038/s41477-019-0547-0