What We’re Reading: November 10th
Review: Growth-mediated plant movements: hidden in plain sight ($)
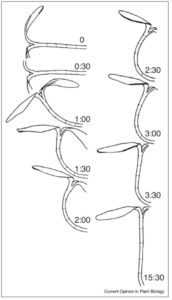 Time-lapse imaging reveals the slow movements of plants, such as phototropism and gravitropism. Harmer and Brooks review the molecular bases for these growth-mediated movements. While auxin has long been known to be involved in photo- and gravitropisms, new results show that ABA is involved in the movement of plants towards water (hydrotropism). The authors also describe autostraightening (realigning to the vertical after tropic bending), which is less well understood but involves proprioception and may involve mechanosensing and the actin/myosin cytoskeleton. (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.10.003
Time-lapse imaging reveals the slow movements of plants, such as phototropism and gravitropism. Harmer and Brooks review the molecular bases for these growth-mediated movements. While auxin has long been known to be involved in photo- and gravitropisms, new results show that ABA is involved in the movement of plants towards water (hydrotropism). The authors also describe autostraightening (realigning to the vertical after tropic bending), which is less well understood but involves proprioception and may involve mechanosensing and the actin/myosin cytoskeleton. (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.10.003
Review: Autophagy as a mediator of life and death in plants ($)
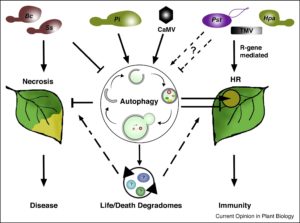 Autophagy is a major pathway involved in degradation and recycling of the cytoplasmic components in a cell. This pathway is functionally well conserved in maintaining cellular homeostasis and modulation of stress responses among yeast, plants and animals. Recent evidence suggests that autophagy targets a range of components in cell machinery. In plants, the role of autophagy in regulating programmed cell death (PCD) remains elusive and hence is a topic of intense investigation. Üstün et al. summarize the latest advances in understanding the mechanism of plant autophagy and discuss the implications of this process in regulating life and death decisions during development and pathogen challenge. The authors highlight selective autophagy mechanisms and shed light on the dual role of this process in either suppression or promotion of developmental, immunity-related and disease-related cell death pathways. The 2016 Nobel Prize for work on autophagy mechanisms should encourage further research in this area and deepen our understanding of these ‘life/death degradomes’. (Summary by Amey Redkar) Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.08.011
Autophagy is a major pathway involved in degradation and recycling of the cytoplasmic components in a cell. This pathway is functionally well conserved in maintaining cellular homeostasis and modulation of stress responses among yeast, plants and animals. Recent evidence suggests that autophagy targets a range of components in cell machinery. In plants, the role of autophagy in regulating programmed cell death (PCD) remains elusive and hence is a topic of intense investigation. Üstün et al. summarize the latest advances in understanding the mechanism of plant autophagy and discuss the implications of this process in regulating life and death decisions during development and pathogen challenge. The authors highlight selective autophagy mechanisms and shed light on the dual role of this process in either suppression or promotion of developmental, immunity-related and disease-related cell death pathways. The 2016 Nobel Prize for work on autophagy mechanisms should encourage further research in this area and deepen our understanding of these ‘life/death degradomes’. (Summary by Amey Redkar) Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.08.011
Inhibition of RNA polymerase II allows controlled mobilization of retrotransposons for plant breeding
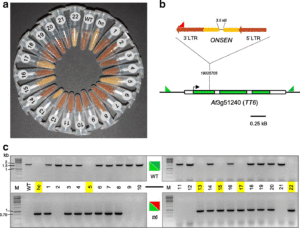 The lack of acceptance of GM-breeding calls for alternative strategies to develop new crop varieties to feed the world’s growing population. Moreover, the regulation of novel approaches for genome editing (CRISPR, TALEN) is still unclear and will potentially remain so for the near future (or will likely be very tight in Europe and parts of Asia and Africa). In this light, the findings of Thieme et al. could offer a new approach to develop genetic diversity in crop plants. By exogenously repressing RNA polymerase II , they showed an increased activity of certain types of transposons that “re-shuffle” the plant’s genome and thus create new genetic and epigenetic diversity. (Summary by Dominik Klauser) Genome Biol. 10.1186/s13059-017-1265-4
The lack of acceptance of GM-breeding calls for alternative strategies to develop new crop varieties to feed the world’s growing population. Moreover, the regulation of novel approaches for genome editing (CRISPR, TALEN) is still unclear and will potentially remain so for the near future (or will likely be very tight in Europe and parts of Asia and Africa). In this light, the findings of Thieme et al. could offer a new approach to develop genetic diversity in crop plants. By exogenously repressing RNA polymerase II , they showed an increased activity of certain types of transposons that “re-shuffle” the plant’s genome and thus create new genetic and epigenetic diversity. (Summary by Dominik Klauser) Genome Biol. 10.1186/s13059-017-1265-4
Recombination between members of Onsen family
 Plant genomes are largely remnants of transposons of varying ages, some of which are presumed to be no longer capable of transposition. Sanchez et al. examined the family of Onsen retrotransposons to determine which members retain activity. They prompted transposition through heat treatment in a mutant line deficient in siRNA production (siRNAs suppress transposon activation). Interestingly, they observed that upon activation the family members recombined to produce new recombination products; “each transposition burst generates a novel progeny population of chromosomally integrated LTR retrotransposons consisting of pairwise recombination products produced in a process comparable the sexual exchange of genetic information.” The outcome is an increase in retrotransposon diversity, similar to what happens during sexual recombination. These findings explain the previously observed high rates of sequence diversification in retrotransposons. (Summary by Mary Williams) Nature Comms. 10.1038/s41467-017-01374-x
Plant genomes are largely remnants of transposons of varying ages, some of which are presumed to be no longer capable of transposition. Sanchez et al. examined the family of Onsen retrotransposons to determine which members retain activity. They prompted transposition through heat treatment in a mutant line deficient in siRNA production (siRNAs suppress transposon activation). Interestingly, they observed that upon activation the family members recombined to produce new recombination products; “each transposition burst generates a novel progeny population of chromosomally integrated LTR retrotransposons consisting of pairwise recombination products produced in a process comparable the sexual exchange of genetic information.” The outcome is an increase in retrotransposon diversity, similar to what happens during sexual recombination. These findings explain the previously observed high rates of sequence diversification in retrotransposons. (Summary by Mary Williams) Nature Comms. 10.1038/s41467-017-01374-x
Amplification of plant disease-resistance genes in pepper is intimately linked to transposon activity
 Peppers are an economically and ecologically important crop plants, but genomic resources are rather scarce. The authors provide here new reference genome sequences for three species of hot pepper (Capsicum baccatum, C. chinense and C. annuum), identifying evolutionary forces that have shaped pepper genomes. Based on these new resources, the study reveals important species-specific chromosomal rearrangements, but also massive amplification of retrotransposons and specific gene families including the nucleotide-binding and leucine-rich-repeats proteins (NLRs), the major disease-resistance gene families in plants. By comparing proliferation dynamics of NLRs in pepper and other plant genomes, the authors show that a large proportion of the NLRs have been created through retroduplication, a process involving the reverse transcription of the NLRs from their mRNA into a complementary DNA copy, mediated by enzymes encoded by retrotransposons. Altogether, this study stresses out the intimate link between genes and transposons in the evolution of genomes, as well as the necessity of high-quality genome references for in-depth genomic studies. (Summary by Matthias Benoit) Genome Biol. 10.1186/s13059-017-1341-9
Peppers are an economically and ecologically important crop plants, but genomic resources are rather scarce. The authors provide here new reference genome sequences for three species of hot pepper (Capsicum baccatum, C. chinense and C. annuum), identifying evolutionary forces that have shaped pepper genomes. Based on these new resources, the study reveals important species-specific chromosomal rearrangements, but also massive amplification of retrotransposons and specific gene families including the nucleotide-binding and leucine-rich-repeats proteins (NLRs), the major disease-resistance gene families in plants. By comparing proliferation dynamics of NLRs in pepper and other plant genomes, the authors show that a large proportion of the NLRs have been created through retroduplication, a process involving the reverse transcription of the NLRs from their mRNA into a complementary DNA copy, mediated by enzymes encoded by retrotransposons. Altogether, this study stresses out the intimate link between genes and transposons in the evolution of genomes, as well as the necessity of high-quality genome references for in-depth genomic studies. (Summary by Matthias Benoit) Genome Biol. 10.1186/s13059-017-1341-9
Complex evolutionary history and targets of domestication in the cultivated potato
 Potatoes originated in the Andes of southern Peru, and are now the third most important crop for direct human consumption. Hardigan et al. sequenced 67 potato relatives, including South American landraces, North American cultivars and wild-diploid species to learn about the genetics of modern potato’s domestication. They identified genes associated with key domestication events including “enlarged tubers, tuber-specific reduction of harmful glycoalkaloids, adaptation to a long-day photoperiod, and reduced sexual fertility.” They found that potatoes have the highest genetic diversity of any sequenced crop, and suggest that the extensive genetic diversity available in wild populations will be useful for crop improvement. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1714380114
Potatoes originated in the Andes of southern Peru, and are now the third most important crop for direct human consumption. Hardigan et al. sequenced 67 potato relatives, including South American landraces, North American cultivars and wild-diploid species to learn about the genetics of modern potato’s domestication. They identified genes associated with key domestication events including “enlarged tubers, tuber-specific reduction of harmful glycoalkaloids, adaptation to a long-day photoperiod, and reduced sexual fertility.” They found that potatoes have the highest genetic diversity of any sequenced crop, and suggest that the extensive genetic diversity available in wild populations will be useful for crop improvement. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1714380114
How asparagus recently changed its lifestyle from hermaphroditism to distinct males and females
 Sex determination in the animal kingdom has been relatively well studied, with two main systems responsible for the sexes in mammals, insects, birds, reptiles and fish; XY and ZW sex-determination. Although much is still unknown about these systems, with many exceptions being discovered to previously considered rules, there is agreement that both these systems are ancient in metazoan lineages. The plant kingdom is notorious for harbouring exceptions to discoveries made in Animalia. The evolution of dioecious (distinct male and female) plants has occurred multiple times and at different time points in the evolution of the flowering plants, making this group ideal to study the evolution of sex chromosomes. Recently, a study of the garden asparagus genome, one of a few dioecious species in an otherwise hermaphroditic genus, revealed a large segment of the Y-chromosome that is absent from the X-chromosome. Harkess et al. also identified two genes likely involved in sex determination present in this 1-million base region; one responsible for suppressing female development and the other for male functionality. Only 6 percent of angiosperms show a dioecious lifestyle, and one-gene and two-gene hypotheses for the origin of dioecy, and an active Y-chromosome that determines the sex of the individual, exist. The asparagus genome lends support to the two-gene model, with the study showing male to hermaphrodite, and male to female conversions, depending on whether one or both genes are deleted. Thus, unlike in Animalia, the origin of a sex system in asparagus is recent, indicating that there are multiple genetic factors likely responsible for the opposite sexes in flowering plants. (Summary by Danielle Roodt Prinsloo) Nature Comms. 10.1038/s41467-017-01064-8
Sex determination in the animal kingdom has been relatively well studied, with two main systems responsible for the sexes in mammals, insects, birds, reptiles and fish; XY and ZW sex-determination. Although much is still unknown about these systems, with many exceptions being discovered to previously considered rules, there is agreement that both these systems are ancient in metazoan lineages. The plant kingdom is notorious for harbouring exceptions to discoveries made in Animalia. The evolution of dioecious (distinct male and female) plants has occurred multiple times and at different time points in the evolution of the flowering plants, making this group ideal to study the evolution of sex chromosomes. Recently, a study of the garden asparagus genome, one of a few dioecious species in an otherwise hermaphroditic genus, revealed a large segment of the Y-chromosome that is absent from the X-chromosome. Harkess et al. also identified two genes likely involved in sex determination present in this 1-million base region; one responsible for suppressing female development and the other for male functionality. Only 6 percent of angiosperms show a dioecious lifestyle, and one-gene and two-gene hypotheses for the origin of dioecy, and an active Y-chromosome that determines the sex of the individual, exist. The asparagus genome lends support to the two-gene model, with the study showing male to hermaphrodite, and male to female conversions, depending on whether one or both genes are deleted. Thus, unlike in Animalia, the origin of a sex system in asparagus is recent, indicating that there are multiple genetic factors likely responsible for the opposite sexes in flowering plants. (Summary by Danielle Roodt Prinsloo) Nature Comms. 10.1038/s41467-017-01064-8
Canalization of tomato fruit metabolism
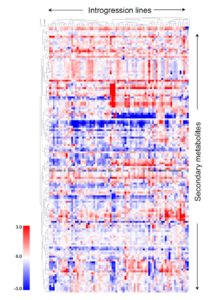 Plants are noted for their phenotypic plasticity, but there are also examples of phenotypic canalization, which Alseekh et al. define as “the property of those phenotypic traits showing no environmental effect when individuals of a specific genotype are exposed to a set of different environments.” In this work, they explored the genetic underpinnings of canalization of tomato fruit metabolism. They examined metabolite profiles from inbred lines taken from three successive harvests to determine the extent of variability of metabolite pools, and identified QTL that are associated with metabolite canalization. The authors observed that, “the intervals they mapped to harbored few metabolism-associated genes, suggesting that the canalization of metabolism is largely controlled by regulatory genes.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00367
Plants are noted for their phenotypic plasticity, but there are also examples of phenotypic canalization, which Alseekh et al. define as “the property of those phenotypic traits showing no environmental effect when individuals of a specific genotype are exposed to a set of different environments.” In this work, they explored the genetic underpinnings of canalization of tomato fruit metabolism. They examined metabolite profiles from inbred lines taken from three successive harvests to determine the extent of variability of metabolite pools, and identified QTL that are associated with metabolite canalization. The authors observed that, “the intervals they mapped to harbored few metabolism-associated genes, suggesting that the canalization of metabolism is largely controlled by regulatory genes.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00367
Mechanochemical polarization of contiguous cell walls shapes plant pavement cells
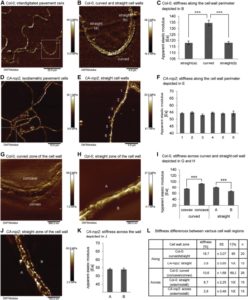
DevCellFig1
The jigsaw-puzzle shape of the epidermis layer has been puzzling the scientists for some time now. Majda et al. examine the shape of the epidermis cells from the cell wall perspective. Mutations leading to even minor changes in cell wall composition significantly affected pavement cell geometry. Using computational modeling the team discovered that in order to produce lobed cells the stiffness of the cell walls must be heterogeneous along and across the cell wall. The heterogeneities in cell wall stiffness were observed in the pavement cells even before lobe formation. The cell wall CA-rop2 mutant, defective in lobe formation, did not show changes in stiffness. Whether mechanochemical polarity exists in other tissues remains to be unveiled. (Summary by Magdalena Julkowska) Devel. Cell 10.1016/j.devcel.2017.10.017
Repression of photomorphogenesis by a small cell-wall-derived dark signal ($)
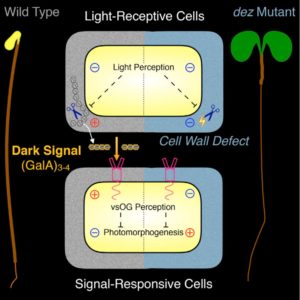 Genetic screens have revealed many key components of light signaling. In this new work, Sinclair et al. provide new insights into the repression of photomorphogenesis by cell-wall derived signals. They started with a mutant, de-etiolated by zinc (dez) that shows open cotyledons and a short hypocotyl (characteristic of photomorphogenesis) in the presence of excess zinc. They mapped the mutation to a gene involved in cell wall modification, specifically pectin acetylation (which can alter the ability of cell walls to bind cations such as Zn2+). The dez mutant phenotype is rescued by the addition of very-short oligogalacturonide; vsOG. The authors propose that in wild-type plants, an extracellular cell-wall derived vsOG signal is produced in the dark through the action of the DEZ protein which supresses photomorphogenesis. The proposed extracellular dark signal could explain the irreversibility and non-cell autonomy of photomorphogenesis. (Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2017.09.063
Genetic screens have revealed many key components of light signaling. In this new work, Sinclair et al. provide new insights into the repression of photomorphogenesis by cell-wall derived signals. They started with a mutant, de-etiolated by zinc (dez) that shows open cotyledons and a short hypocotyl (characteristic of photomorphogenesis) in the presence of excess zinc. They mapped the mutation to a gene involved in cell wall modification, specifically pectin acetylation (which can alter the ability of cell walls to bind cations such as Zn2+). The dez mutant phenotype is rescued by the addition of very-short oligogalacturonide; vsOG. The authors propose that in wild-type plants, an extracellular cell-wall derived vsOG signal is produced in the dark through the action of the DEZ protein which supresses photomorphogenesis. The proposed extracellular dark signal could explain the irreversibility and non-cell autonomy of photomorphogenesis. (Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2017.09.063
Duplication of an upstream silencer of FZP increases grain yield in rice
 The trade-off between grain size and number has been traditionally hard to break. Bai et al. established that differences in panicle architecture between two rice cultivars resulted from duplication of a long-distance silencer of the FRIZZY PANICLE (FZP) gene in one cultivar (Chuan 7). Increased silencing of FZP led to a longer panicle branching period and grain yield increases of at least 15% in Chuan 7. This was due to a large increase in the number of spikelets per panicle, with only a slight reduction in grain weight. Further molecular characterisation revealed that OsBZR1 binds to the silencer sequence to repress FZP transcription. Full FZP knock-outs are not viable, but fine-tuning of FZP expression appears able to enhance rice yields by optimization of the grain size/number trade-off. (Summary by Mike Page) Nature Plants 10.1038/s41477-017-0042-4
The trade-off between grain size and number has been traditionally hard to break. Bai et al. established that differences in panicle architecture between two rice cultivars resulted from duplication of a long-distance silencer of the FRIZZY PANICLE (FZP) gene in one cultivar (Chuan 7). Increased silencing of FZP led to a longer panicle branching period and grain yield increases of at least 15% in Chuan 7. This was due to a large increase in the number of spikelets per panicle, with only a slight reduction in grain weight. Further molecular characterisation revealed that OsBZR1 binds to the silencer sequence to repress FZP transcription. Full FZP knock-outs are not viable, but fine-tuning of FZP expression appears able to enhance rice yields by optimization of the grain size/number trade-off. (Summary by Mike Page) Nature Plants 10.1038/s41477-017-0042-4
Changes in the allocation of endogenous strigolactones improve plant biomass production on phosphate-poor soils
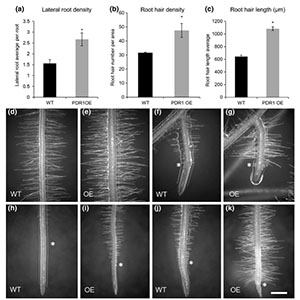 Strigolactones are plant hormones with pivotal roles in plant growth, such as shoot development, root system architecture and plant-microbe interactions. PDR is a strigolactone transporter that was previously identified in Petunia by the Martinoia lab. In this work, Liu et al. show that the overexpression of PDR under the 35SCaMV promoter dramatically improves Petunia fitness in different kinds of low phosphate soils. PDR overexpression caused strigolactone mislocalization, inducing a wide spectrum of phenotypic changes such as increased root apparatus, longer root hairs, bigger biomass and higher mycorrhizal colonization. Therefore a manipulation of the strigolactone pathway represents a viable strategy for improving plant growth in low-phosphorus environments. (Summary by Marco Giovanetti) New Phytol. 10.1111/nph.14847
Strigolactones are plant hormones with pivotal roles in plant growth, such as shoot development, root system architecture and plant-microbe interactions. PDR is a strigolactone transporter that was previously identified in Petunia by the Martinoia lab. In this work, Liu et al. show that the overexpression of PDR under the 35SCaMV promoter dramatically improves Petunia fitness in different kinds of low phosphate soils. PDR overexpression caused strigolactone mislocalization, inducing a wide spectrum of phenotypic changes such as increased root apparatus, longer root hairs, bigger biomass and higher mycorrhizal colonization. Therefore a manipulation of the strigolactone pathway represents a viable strategy for improving plant growth in low-phosphorus environments. (Summary by Marco Giovanetti) New Phytol. 10.1111/nph.14847
Roots-eye view: Using microdialysis and microCT to non-destructively map root nutrient depletion and accumulation zones
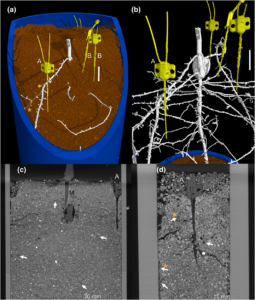 Plant roots constantly engage in nutrient and water uptake for crop productivity. Increasing the nutrient uptake efficiency of roots will promote sustainable agriculture by decreasing the need for fertilizer applications. To achieve this task, we need to understand the physiology of intact roots in their natural soil environment. Brackin et al. examined root nutrient foraging ability by using microdialysis to measure ion concentrations and microCT to image roots. By using probes placed adjacent to the roots in a non destructive manner, the authors were able to measure nitrate and ammonium levels in the rhizosphere in the vicinity of roots. The authors observed that close to roots there is an accumulation of nitrate but a depletion of ammonium, as a consequences of the difference in mobility of the two ions in the soil. This combination of complementary techniques could be helpful in advancing our understanding of the allocation of nutrients in the roots across varied environmental conditions, and allow an efficient means of selecting crops with increased nutrient efficiency to promote sustainable agriculture. (Summary by Amey Redkar) Plant Cell Environ. 10.1111/pce.13072/full
Plant roots constantly engage in nutrient and water uptake for crop productivity. Increasing the nutrient uptake efficiency of roots will promote sustainable agriculture by decreasing the need for fertilizer applications. To achieve this task, we need to understand the physiology of intact roots in their natural soil environment. Brackin et al. examined root nutrient foraging ability by using microdialysis to measure ion concentrations and microCT to image roots. By using probes placed adjacent to the roots in a non destructive manner, the authors were able to measure nitrate and ammonium levels in the rhizosphere in the vicinity of roots. The authors observed that close to roots there is an accumulation of nitrate but a depletion of ammonium, as a consequences of the difference in mobility of the two ions in the soil. This combination of complementary techniques could be helpful in advancing our understanding of the allocation of nutrients in the roots across varied environmental conditions, and allow an efficient means of selecting crops with increased nutrient efficiency to promote sustainable agriculture. (Summary by Amey Redkar) Plant Cell Environ. 10.1111/pce.13072/full
Modeling guard cell-to-leaf scales with OnGuard2
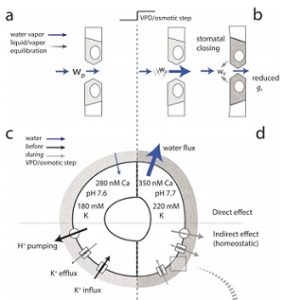 While much is known about the processes involved in stomatal movement and the processes involved in the transpiration of leaves, there has been no framework to bridge this micro-macro divide. Wang and colleagues bridge this divide through OnGuard2, a quantitative systems platform that uses the molecular mechanisms of the guard cell (ion transport, metabolism, signaling) to understand the transpiration of the leaf. They show that OnGuard2 reproduces experimentally-derived foliar transpiration and stomatal kinetics of different Arabidopsis lines, including wild-type, slac1 Cl– channel mutant and ost2 H+-ATPase mutant lines. This single framework can be used to understand stomatal physiology in greater detail, and can be used to connect guard cell membrane transport to plant water relations. (Summary by Julia Miller) Plant Cell 10.1105/tpc.17.00694
While much is known about the processes involved in stomatal movement and the processes involved in the transpiration of leaves, there has been no framework to bridge this micro-macro divide. Wang and colleagues bridge this divide through OnGuard2, a quantitative systems platform that uses the molecular mechanisms of the guard cell (ion transport, metabolism, signaling) to understand the transpiration of the leaf. They show that OnGuard2 reproduces experimentally-derived foliar transpiration and stomatal kinetics of different Arabidopsis lines, including wild-type, slac1 Cl– channel mutant and ost2 H+-ATPase mutant lines. This single framework can be used to understand stomatal physiology in greater detail, and can be used to connect guard cell membrane transport to plant water relations. (Summary by Julia Miller) Plant Cell 10.1105/tpc.17.00694
CIRCADIAN CLOCK ASSOICATED 1 (CCA1) and the circadian control of stomatal aperture
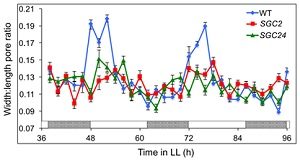 How do plants ‘decide’ how to respond to so many conflicting stimuli? What has the final say in stomatal aperture control? Hassidium et al. investigate the role of the oscillator gene CIRCADIAN CLOCK ASSOICATED 1 (CCA1) during stomatal opening and determined if CCA1 is responsive to other stimuli. Overexpression of CCA1 specifically in guard cells (SGC plants) results in higher CCA1 levels that are arrhythmic in nature and display impaired opening and closing in response to light. However, SGC guard cells responded normally to opening buffer and ABA. By analyzing expression levels of photoperiod pathway genes, the authors found an overlap between the FLOWERING LOCUS T (FT) and CCA1 pathway to regulate guard cell apertures. SGC plants showed decreased levels of FT, which was previously shown to positively regulate stomatal opening. Lastly, differences between WT guard cell dynamics grown in long day versus short day were observed: short day plants had lower FT levels and impaired stomatal dynamics. These data indicate that the circadian clock in concert with the photoperiod pathway is involved in regulating stomatal apertures. Understanding the mechanism of how different stimuli control stomatal dynamics will not only further our understanding of stomatal dynamics but also how plants respond to varying external and internal stimuli. (Summary by Alecia Biel) Plant Phys. 10.1104/pp.17.01214.
How do plants ‘decide’ how to respond to so many conflicting stimuli? What has the final say in stomatal aperture control? Hassidium et al. investigate the role of the oscillator gene CIRCADIAN CLOCK ASSOICATED 1 (CCA1) during stomatal opening and determined if CCA1 is responsive to other stimuli. Overexpression of CCA1 specifically in guard cells (SGC plants) results in higher CCA1 levels that are arrhythmic in nature and display impaired opening and closing in response to light. However, SGC guard cells responded normally to opening buffer and ABA. By analyzing expression levels of photoperiod pathway genes, the authors found an overlap between the FLOWERING LOCUS T (FT) and CCA1 pathway to regulate guard cell apertures. SGC plants showed decreased levels of FT, which was previously shown to positively regulate stomatal opening. Lastly, differences between WT guard cell dynamics grown in long day versus short day were observed: short day plants had lower FT levels and impaired stomatal dynamics. These data indicate that the circadian clock in concert with the photoperiod pathway is involved in regulating stomatal apertures. Understanding the mechanism of how different stimuli control stomatal dynamics will not only further our understanding of stomatal dynamics but also how plants respond to varying external and internal stimuli. (Summary by Alecia Biel) Plant Phys. 10.1104/pp.17.01214.
Measures of interoperability of phenotypic data: minimum information requirements and formatting

PlantMethFig1
If you ever tried to compare the results from two different experiments, you probably realized that small variations in the environment have big impact on how the plants grow. There is a lot to be learned from how the variation in the environment affects plant phenotypes, but currently the description of experimental setup varies between studies. Cwiek-Kupczynska et al. propose Minimum Information About a Plant Phenotyping Experiment (MIAPPE) aiming to unify experimental setup details provided alongside the experiment results. Application of MIAPPE format will not only ensure FAIR data (Findable, Accessible, Interoperable and Reproducible), but will allow us to learn more about genotype-environment interactions by comparing the phenotypic data on another, bigger scale. (Summary by Magdalena Julkowska) Plant Methods 10.1186/s13007-016-0144-4



