What We’re Reading: July 7
 This week’s edition of What We’re Reading is guest edited by K.K. Sabu. Sabu is educated in the field of Plant Sciences and has working experience in plant genetics and genomics since 1995. His research interests include response of plants against environmental changes, natural variations in plant genetic resources and incorporation of genomics tools for big data analysis. He is supervising students for their MSc, MPhil and PhD programme and carrying out externally funded research programmes.
This week’s edition of What We’re Reading is guest edited by K.K. Sabu. Sabu is educated in the field of Plant Sciences and has working experience in plant genetics and genomics since 1995. His research interests include response of plants against environmental changes, natural variations in plant genetic resources and incorporation of genomics tools for big data analysis. He is supervising students for their MSc, MPhil and PhD programme and carrying out externally funded research programmes.
Review: Exocytic trafficking pathways in plants
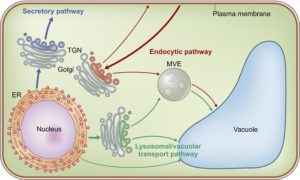 The membrane trafficking system plays an important role in transporting various substances, including proteins, lipids, and small molecules throughout cells. Secretory/exocytic trafficking pathways are involved in various biological functions, including development, plant–microbe interactions and cell-to-cell communications. This review by Kanazawa and Ueda describes plant-unique functions involving the exocytic pathways that may have resulted from their unique diversification during land plant evolution. New Phytol. 10.1111/nph.14613
The membrane trafficking system plays an important role in transporting various substances, including proteins, lipids, and small molecules throughout cells. Secretory/exocytic trafficking pathways are involved in various biological functions, including development, plant–microbe interactions and cell-to-cell communications. This review by Kanazawa and Ueda describes plant-unique functions involving the exocytic pathways that may have resulted from their unique diversification during land plant evolution. New Phytol. 10.1111/nph.14613
Review: Ecology and genomic insights on plant endophytes ($)
 Plants live in a microbe-rich world. Many microorganisms colonize the surfaces of plants, whereas endophytes live within plant tissues. Although some endophytes do not seem to have positive or negative effects on the host plant, others are mutualists (forming mutually beneficial relationship) or harmful pathogens. The nature of these interactions is not fixed, but depends a complex network that involves abiotic factors and the diversity and abundance of other microorganism. Brader et al. discuss the factors that underlie different plant-endophyte interactions in terms of host plant response, colonization strategy, genome content and abiotic and biotic factors. Annu. Rev. Phytopathol. 10.1146/annurev-phyto-080516-035641
Plants live in a microbe-rich world. Many microorganisms colonize the surfaces of plants, whereas endophytes live within plant tissues. Although some endophytes do not seem to have positive or negative effects on the host plant, others are mutualists (forming mutually beneficial relationship) or harmful pathogens. The nature of these interactions is not fixed, but depends a complex network that involves abiotic factors and the diversity and abundance of other microorganism. Brader et al. discuss the factors that underlie different plant-endophyte interactions in terms of host plant response, colonization strategy, genome content and abiotic and biotic factors. Annu. Rev. Phytopathol. 10.1146/annurev-phyto-080516-035641
Review: The algal revolution ($)
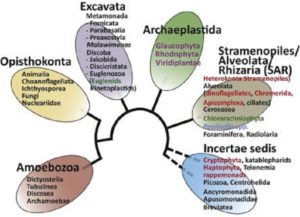 Algae are eukaryotes and they are mostly photosynthetic. These life forms occupy multiple branches of the tree of life, and are vital for planet function and health, yet only about 10% of algal species have been described. Brodie et al. review new insights and applications, boosted by high-throughput –omics approaches, for this important group of organsisms. Special sections focus on the potential of algae in biotechnology for high-value industrial products and food supplements, the evolutionary origins of algae and algal mulitcelluarity, and the application of new tools in algal science. This review also looks sex determination in algae and how these studies inform our understanding of the evolution of sex chromosomes. Trends Plant Sci. 10.1016/j.tplants.2017.05.005
Algae are eukaryotes and they are mostly photosynthetic. These life forms occupy multiple branches of the tree of life, and are vital for planet function and health, yet only about 10% of algal species have been described. Brodie et al. review new insights and applications, boosted by high-throughput –omics approaches, for this important group of organsisms. Special sections focus on the potential of algae in biotechnology for high-value industrial products and food supplements, the evolutionary origins of algae and algal mulitcelluarity, and the application of new tools in algal science. This review also looks sex determination in algae and how these studies inform our understanding of the evolution of sex chromosomes. Trends Plant Sci. 10.1016/j.tplants.2017.05.005
Perspective: Generating virtual crops using in silico modeling platform
 “We are at a point in history where we have both the need and the capability to use information and models at multiple levels to model whole systems and to achieve greater insights into how whole plants and ecosystems will respond to genetic changes, as well as environmental challenges never before encountered,” state Marshall-Colon et al., the authors of this persective. They describe how in silico multi-scale modelling tools that link gene networks, protein synthesis, metabolic pathways, physiology, and growth, integrated with ecosystem, weather, and climate models, help to predict how various interactions respond to environmental perturbations. These models have the ability to fill in missing mechanistic details and propose new hypotheses to prioritize directed engineering efforts resulting in the improvement of crop yield, sustainability, and increase future food security. The authors propose “the creation of a discovery platform called Crops in silico (Cis)” and lay out a framework to achieve these objectives. Front. Plant Sci. 10.3389/fpls.2017.00786
“We are at a point in history where we have both the need and the capability to use information and models at multiple levels to model whole systems and to achieve greater insights into how whole plants and ecosystems will respond to genetic changes, as well as environmental challenges never before encountered,” state Marshall-Colon et al., the authors of this persective. They describe how in silico multi-scale modelling tools that link gene networks, protein synthesis, metabolic pathways, physiology, and growth, integrated with ecosystem, weather, and climate models, help to predict how various interactions respond to environmental perturbations. These models have the ability to fill in missing mechanistic details and propose new hypotheses to prioritize directed engineering efforts resulting in the improvement of crop yield, sustainability, and increase future food security. The authors propose “the creation of a discovery platform called Crops in silico (Cis)” and lay out a framework to achieve these objectives. Front. Plant Sci. 10.3389/fpls.2017.00786
Day length−dependent regulation of rice flowering ($)
 Flowering in rice (Oryza sativa) is regulated by photoperiodicity, with Heading date 1 (Hd1) having a pivotal role. It promotes flowering under short-day conditions (SD) and represses it under long-day conditions (LD), however, the molecular mechanism of how it works remains largely unknown. Du et al. show that the repression of flowering in LD by Hd1 is dependent on the transcription factor DAYS TO HEADING 8 (DTH8). Through biochemical and genetic approaches, the authors show that DTH8 works in concert with Hd1 to repress expression of another gene, Hd3a, through H3K27 trimethylation (H3K27me3), thus revealing a new insight into photoperiodic control of flowering. Mol. Plant 10.1016/j.molp.2017.05.006
Flowering in rice (Oryza sativa) is regulated by photoperiodicity, with Heading date 1 (Hd1) having a pivotal role. It promotes flowering under short-day conditions (SD) and represses it under long-day conditions (LD), however, the molecular mechanism of how it works remains largely unknown. Du et al. show that the repression of flowering in LD by Hd1 is dependent on the transcription factor DAYS TO HEADING 8 (DTH8). Through biochemical and genetic approaches, the authors show that DTH8 works in concert with Hd1 to repress expression of another gene, Hd3a, through H3K27 trimethylation (H3K27me3), thus revealing a new insight into photoperiodic control of flowering. Mol. Plant 10.1016/j.molp.2017.05.006
Diversity and distribution of seed physical dormancy
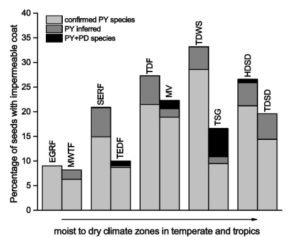 Physical dormancy (PY) as a consequence of an impermeable seed or fruit coat occurs only in several genera of 18 angiosperm families and plays an important role in seed germination. It has been speculated that PY is more prevalent in dry lands compared to moist vegetation zones. Using a data set of 13,792 species, Jaganathan et al. found that the number of species with PY in tropics (19%) is higher than the number of PY species in the temperate ecosystem (15%). However, in both tropic and temperate regions, PY is less common in moist and low-temperature vegetation zones compared with dry and high-temperature vegetation. The paper discusses the ecological adaptation of PY in the dry ecosystem and considers the mechanism of persistence and dormancy break in PY and physiological dormant (PD) species. Plant Science Today 10.14719/pst.2017.4.2.293
Physical dormancy (PY) as a consequence of an impermeable seed or fruit coat occurs only in several genera of 18 angiosperm families and plays an important role in seed germination. It has been speculated that PY is more prevalent in dry lands compared to moist vegetation zones. Using a data set of 13,792 species, Jaganathan et al. found that the number of species with PY in tropics (19%) is higher than the number of PY species in the temperate ecosystem (15%). However, in both tropic and temperate regions, PY is less common in moist and low-temperature vegetation zones compared with dry and high-temperature vegetation. The paper discusses the ecological adaptation of PY in the dry ecosystem and considers the mechanism of persistence and dormancy break in PY and physiological dormant (PD) species. Plant Science Today 10.14719/pst.2017.4.2.293
Abiotic and biotic stress adaptation in Arabidopsis ($)
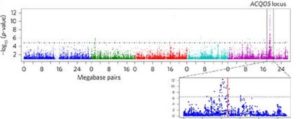 Drought, salt or cold stress often decreases plant fitness. Ariga et al. show that the ability of plants to withstand the stress conditions is controlled by certain genes such as ACQUIRED OSMOTOLERANCE (ACQOS), which encodes a nucleotide-binding leucine-rich repeat (NLR) protein. Natural variations at the ACQOS locus suggest that there are functional and non-functional ACQOS that are being maintained due to a trade-off between biotic and abiotic stress adaptation. Thus, it could be inferred that competing environmental stresses influence polymorphism in certain plant NLR genes. Nature Plants 10.1038/nplants.2017.72
Drought, salt or cold stress often decreases plant fitness. Ariga et al. show that the ability of plants to withstand the stress conditions is controlled by certain genes such as ACQUIRED OSMOTOLERANCE (ACQOS), which encodes a nucleotide-binding leucine-rich repeat (NLR) protein. Natural variations at the ACQOS locus suggest that there are functional and non-functional ACQOS that are being maintained due to a trade-off between biotic and abiotic stress adaptation. Thus, it could be inferred that competing environmental stresses influence polymorphism in certain plant NLR genes. Nature Plants 10.1038/nplants.2017.72




Leave a Reply
Want to join the discussion?Feel free to contribute!