Unregulated Sphingolipid Biosynthesis in Arabidopsis ORM Mutants
Gonzalez Solis et al. describe the metabolic, cellular, and physiological consequences of the complete loss of regulation of sphingolipid biosynthesis. Plant Cell https://doi.org/10.1105/tpc.20.00015
By Ariadna Gonzalez Solis and Edgar B. Cahoon
Background: Glycosphingolipids are molecules with backbones derived from serine and palmitoyl-CoA that are essential for endomembrane and plasma membrane function. The sphingolipid biosynthetic intermediates long chain bases (LCBs) and ceramides also contribute to signaling processes associated with abiotic stress responses and programmed cell death (PCD) induction during pathogenesis. Orosomucoid-like proteins (ORMs) negatively regulate sphingolipid biosynthesis, a reversible process critical for balancing the sphingolipid levels needed for plant growth and PCD induction. ORM proteins physically interact with serine palmitoyltransferase (SPT), the first enzyme of LCB biosynthesis, to help maintain sphingolipid homeostasis. However, the molecular basis for this interaction and its metabolic and physiological significance are not well understood.
Question: What are the physiological consequences of unregulated sphingolipid biosynthesis in an Arabidopsis ORM double knockout mutant? What are the molecular mechanisms associated with ORM-mediated SPT regulation?
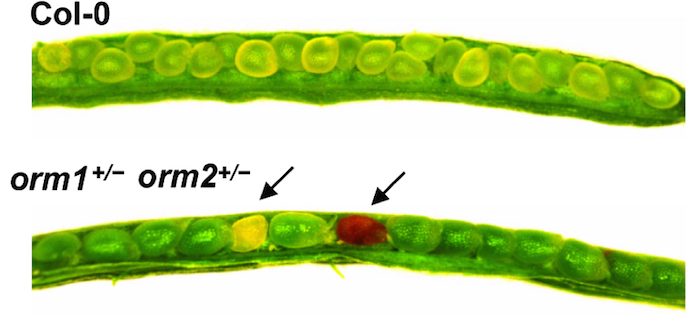
Findings: Using complete knockout mutants for the two Arabidopsis thaliana ORM genes (ORM1 and ORM2) obtained by gene editing, we discovered that removing sphingolipid biosynthetic regulation results in nonviable seeds with impaired embryo development, ceramide hyperaccumulation, and strongly reduced oil content. We obtained identical phenotypes by removing the first transmembrane domain of the LCB1 subunit SPT, which is known to be essential for the ORM-SPT interaction. We also identified a mutant with a single amino acid deletion of ORM1 in an ORM2 knockout background that did not survive beyond the seedling stage. This mutant accumulated ceramides, LCBs, and atypical sphingolipids. In addition, this mutant had severely altered organellar structures and induced expression of sphingolipid catabolic and pathogenesis-related genes. Using a yeast model, we showed that ORM1 with one amino acid deletion strongly reduced the ORM-LCB1 interaction, providing structural clues about ORM regulation of SPT.
Next steps: We aim to understand how ORM proteins sense intracellular sphingolipid concentrations and reversibly regulate SPT activity for sphingolipid homeostasis to support growth but also to respond to environmental stimuli, such as pathogen attack that induces hypersensitive-response PCD. We are also attempting to elucidate structural aspects of ORM proteins that underlie their interaction with SPT.
Ariadna Gonzalez-Solis, Gongshe Han, Lu Gan, Yunfeng Liu, Rebecca E. Cahoon, Jonathan E. Markham, Teresa M. Dunn, Edgar B. Cahoon (2020). Unregulated Sphingolipid Biosynthesis in Gene-Edited Arabidopsis ORM Mutants Results in Nonviable Seeds with Strongly Reduced Oil Content. Plant Cell DOI: https://doi.org/10.1105/tpc.20.00015.