TPLATE complex subunits are recruited simultaneously to the plasma membrane
Kerri Hunter
Viikki Plant Science Centre, Organismal and Evolutionary Biology Research Programme, Faculty of Biological and Environmental Sciences, University of Helsinki, Helsinki, Finland
ORCID: 0000-0002-2285-6999
Endocytosis is the process by which extracellular material and plasma membrane (PM) components are internalized by cells. Clathrin-mediated endocytosis, the most common form of endocytosis in eukaryotes, is required for a multitude of cellular processes, and is essential for proper cell-to-cell communication and responses to environmental stimuli (Gadeyne et al. 2014). However, clathrin itself does not bind directly to the PM or cargo proteins, and thus adapter protein complexes are required to fulfil the important role of linking the PM and the clathrin cage (McMahon & Boucrot, 2011). One of the two main adaptor protein complexes found in plants is the TPLATE complex (TPC). The TPC is an evolutionary ancient complex that has been retained in plants (Hirst et al., 2014), and is recruited to the PM during the early stages of endocytosis (Gadeyne et al. 2014; Narasimhan et al., 2020). It has been identified at the PM as an octomeric complex containing the six-subunit TPC core, and two plant-specific subunits, AtEH1/Pan1 and AtEH2/Pan1 (Gadeyne et al., 2014; Hirst et al., 2014). Whereas a relationship between TPC subunit composition and localization has been observed (Gaydene et al., 2014), the dynamics of TPC subunit recruitment and assembly at the PM have not been previously defined in plants.
Much of the current knowledge on clathrin-mediated endocytosis comes from mammalian and yeast cells, whereas endocytic processes in plants remain less well characterized. This is due in part to the differential dynamics and time scale of endocytic events among different organisms. With the exception of neurons, endocytosis occurs on a time scale of minutes in mammalian cells and yeast (Balaji and Ryan, 2007; Lu et al., 2016; Taylor et al., 2011). In plants, the time scale is much faster, and larger clathrin cages can be formed in shorter time periods (Narasimhan et al., 2020). These faster dynamics present a technical challenge in plants to observe and monitor the recruitment of proteins and the precise sequence of events involved (Zhang et al., 2015). New methods are therefore needed to overcome these challenges and enhance the temporal visualization of the cellular dynamics directing endocytosis.
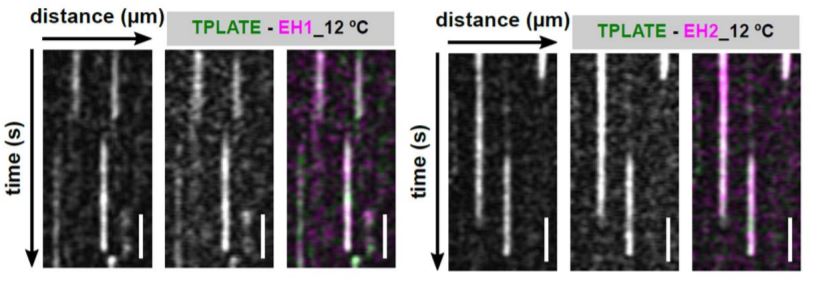
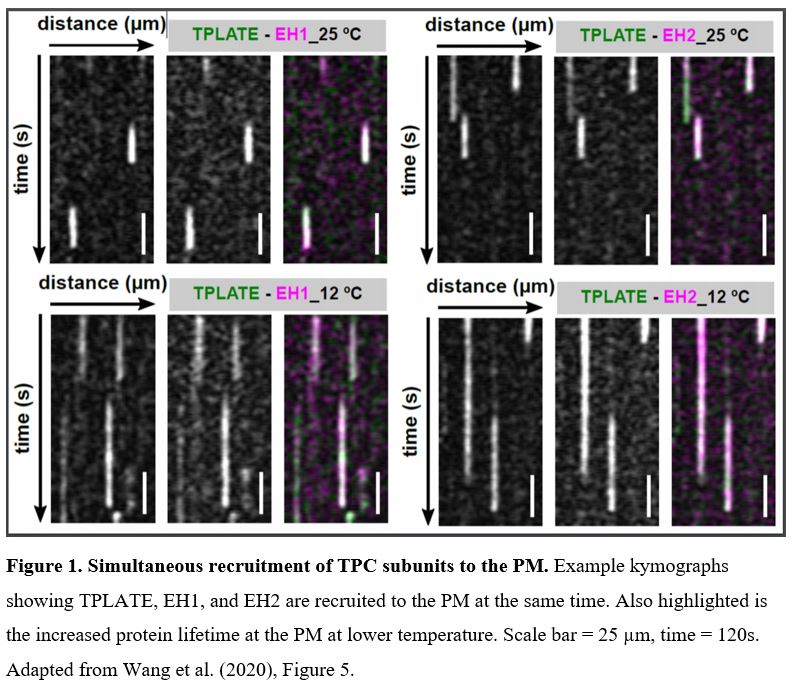 In this issue of Plant Physiology, Wang et al. (2020) utilized an innovative approach to increase the temporal resolution for live-cell imaging of endocytic events by lowering the sample temperature. Their microfluidics-based method enabled rapid, on-slide manipulation of sample temperature. The authors first demonstrated that lowering the sample temperature from 25°C to 12°C effectively slows the cellular processes involved in clathrin-mediated endocytosis, prolonging the lifetime of endocytic proteins at the PM and allowing for enhanced temporal resolution during imaging (Wang et al., 2020). Utilizing advanced time-lapse imaging and kymograph analysis, the authors show that the TPLATE, AtEH/Pan1, and AtEH/Pan1 TPC subunits are recruited to the PM simultaneously, at both 25°C and 12°C (Wang et. al., 2020). The data presented by Wang et al. (2020) suggest that the TPC arrives at the PM as an octomeric complex, rather than via sequential recruitment of the comprising subunits. This insight into the assembly of the TPC at the PM, and the roles of the TPC subunits in attaining proper subcellular localization, serves to further the understanding of how clathrin-mediated endocytosis progresses in plants.
In this issue of Plant Physiology, Wang et al. (2020) utilized an innovative approach to increase the temporal resolution for live-cell imaging of endocytic events by lowering the sample temperature. Their microfluidics-based method enabled rapid, on-slide manipulation of sample temperature. The authors first demonstrated that lowering the sample temperature from 25°C to 12°C effectively slows the cellular processes involved in clathrin-mediated endocytosis, prolonging the lifetime of endocytic proteins at the PM and allowing for enhanced temporal resolution during imaging (Wang et al., 2020). Utilizing advanced time-lapse imaging and kymograph analysis, the authors show that the TPLATE, AtEH/Pan1, and AtEH/Pan1 TPC subunits are recruited to the PM simultaneously, at both 25°C and 12°C (Wang et. al., 2020). The data presented by Wang et al. (2020) suggest that the TPC arrives at the PM as an octomeric complex, rather than via sequential recruitment of the comprising subunits. This insight into the assembly of the TPC at the PM, and the roles of the TPC subunits in attaining proper subcellular localization, serves to further the understanding of how clathrin-mediated endocytosis progresses in plants.
Using on-slide sample cooling as a means to slow cellular processes is a promising approach to increase the spatiotemporal resolution of time-lapse imaging experiments. It is however, as the authors note, important to ensure that the temperature changes do not significantly alter the process itself under investigation. Deviations from the normal growth temperature can influence membrane composition and fluidity, as well as cytoskeleton dynamics. Nevertheless, Wang et al. (2020) have convincingly demonstrated that the observed simultaneous recruitment of TPLATE, AtEH1/Pan1, and AtEH2/Pan1 is consistent at both physiological and lowered temperatures. Thus, this method of lowering the temperature at which imaging is performed can be a valuable tool for improved visualization of other fast-acting cellular events or protein interactions.
References:
Balaji, J., and Ryan, T.A. (2007). Single-vesicle imaging reveals that synaptic vesicle exocytosis and endocytosis are coupled by a single stochastic mode. Proc Natl Acad Sci U S A 104, 20576-20581.
Gadeyne, A., Sanchez-Rodriguez, C., Vanneste, S., Di Rubbo, S., Zauber, H., Vanneste, K., Van Leene, J., De Winne, N., Eeckhout, D., Persiau, G., et al. (2014). The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156, 691-704.
Hirst, J., Schlacht, A., Norcott, J.P., Traynor, D., Bloomfield, G., Antrobus, R., Kay, R.R., Dacks, J.B., and Robinson, M.S. (2014). Characterization of TSET, an ancient and widespread membrane trafficking complex. Elife 3, e02866.
Lu, R., Drubin, D.G., and Sun, Y. (2016). Clathrin-mediated endocytosis in budding yeast at a glance. J Cell Sci 129, 1531-1536.
McMahon, H.T., and Boucrot, E. (2011). Molecular mechanism and physiological functions of clathrin mediated endocytosis. Nat Rev Mol Cell Biol 12, 517-533.
Narasimhan, M., Johnson, A., Prizak, R., Kaufmann, W.A., Tan, S., Casillas-Perez, B., and Friml, J. (2020). Evolutionarily unique mechanistic framework of clathrin-mediated endocytosis in plants. Elife 9.
Taylor, M.J., Perrais, D., and Merrifield, C.J. (2011). A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol 9, e1000604.
Zhang, Y., Persson, S., Hirst, J., Robinson, M.S., van Damme, D., and Sanchez-Rodriguez, C. (2015). Change your TPLATE, change your fate: plant CME and beyond. Trends Plant Sci 20, 41-48.