A damascene moment: the genetic basis of complex petals in Nigella
From the flat white sheets of our favourite weed Arabidopsis thaliana to the colourful cups and spirals of orchids, petals come in a spectacular array of shapes, sizes, colours and textures. These elaborate forms have often evolved to attract pollinators. For example, bee orchids produce petals that mimic female bees to attract amorous males for pollination. Thousands of species have similarly elaborate petals with teeth, spurs or pockets that entice pollinators (Endress and Matthews, 2006). However, much of our knowledge of petal development comes from studies in plants with simple petals such as A. thaliana. How this basic developmental program has been modified to produce more complex structures is still poorly understood. This is in part because it has been difficult to find species with complex petals that are genetically and developmentally tractable.
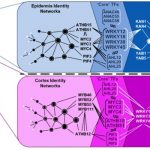
 In this issue of The Plant Cell, Zhang et al. (2020) begin to address this problem by combining transcriptomic and functional analyses in Nigella damascena to understand the genetic basis of elaborate petal formation. Their work not only provides a broad overview of gene expression throughout complex petal development, but also sets the stage for detailed functional analysis of individual candidates and illustrates the potential of using non-model plants to investigate questions of fundamental importance.
In this issue of The Plant Cell, Zhang et al. (2020) begin to address this problem by combining transcriptomic and functional analyses in Nigella damascena to understand the genetic basis of elaborate petal formation. Their work not only provides a broad overview of gene expression throughout complex petal development, but also sets the stage for detailed functional analysis of individual candidates and illustrates the potential of using non-model plants to investigate questions of fundamental importance.
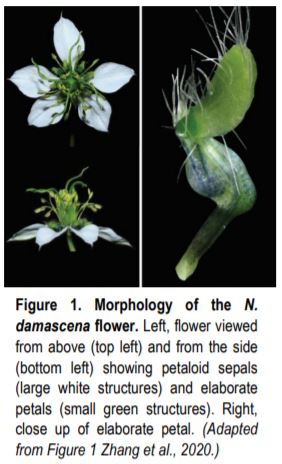
Along with complex petals, N. damascena produces simple petaloid sepals. This allowed Zhang and colleagues to compare the development of complex and simple forms within the same flower. They began by analysing petal, sepal, stamen, and carpel transcriptomes throughout development and showed that at early stages all organs express similar genes and as development progresses, different organ types build up more and more expression differences. They identified 918 genes specifically expressed in petals and another 1,214 genes upregulated in petals. These include genes already known to function in N. damascena petal development such as homologues of the floral regulator APETELA3 (AP3) (Gonçalves et al. 2013) and genes previously unknown in petal development, such as homologues of LATE MERISTEM IDENTITY 1 (LMI1), a regulator of complex leaf formation (Vlad et al. 2014) and several regulators of meristem formation. This data set builds up a molecular picture of petal development and hints that genes usually involved in leaf or meristem development may have been co-opted to produce elaborate petal shapes.
To identify genes that may control petal morphology, the authors analysed the expression of candidate genes from developmental programs that control various aspects of leaf and flower development. They identified 30 candidates expressed at key points of morphological change and suggest that they may control these developmental transitions. These include roles for homologues of CUP SHAPED COTYLEDON 1 (CUC1) in petal bifurcation and LMI1 in short trichome differentiation.
To test if these candidates are involved in petal development, they performed RNA in situ hybridisation and showed that, as predicted, LMI1 is expressed in petals where short trichomes form, but also coincides with the bifurcation of the lower lip. They then used Virus-Induced Gene Silencing (VIGS) to functionally test the role of LMI1 in petal development and showed that when LMI1 expression is knocked down, the lower lip does not bifurcate and short trichomes do not form. These data show that LMI1 contributes to elaborate petal development and suggest that other candidates from the authors’ expression analysis are worth pursuing in future functional studies.
Together these results provide an atlas of gene expression throughout elaborate petal development and identify many candidate genes that may sculpt this complex shape. In particular, the role of LMI1 in petal development reinforces the idea that genes are often co-opted from other developmental programs to evolve novel forms. Future work promises to analyse these candidates in detail to understand whether co-option is the rule for complex petal development, or whether novel genes are also involved. This work highlights the potential of using genetic analysis in non-model species to address scientific questions that are inaccessible with traditional models. This work should encourage functional studies in other non-model systems and bring further developmental and evolutionary questions within reach.
Chris Whitewoods
Department of Cell and Developmental Biology
John Innes Centre, Norwich
ORCID: 0000-0001-6886-3572
REFERENCES
Endress, P.K., and Matthews, M. (2006). Elaborate petals and staminodes in eudicots: Diversity, function, and evolution. Org. Divers.
Evol. 6:257-293.
Gonçalves, B., Nougué, O., Jabbour, F., Ridel, C., Morin, H., Laufs, P., Manicacci, D., and Damerval, C. (2013). An APETALA3 homolog controls both petal identity and floral meristem patterning in Nigella damascena L. (Ranunculaceae). Plant J. 76:223-235.
Vlad, D., Kierzkowski, D., Rast, M.I., Vuolo, F., Dello Ioio, R., Galinha, C., Gan, X., Hajheidari, M., Hay, A., Smith, R.S., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343:780-783.