To Be or Not To Be Pathogenic: Transcriptional Reprogramming Dictates a Fungal Pathogen’s Response to Different Hosts
The fungal pathogen Fusarium virguliforme invades a wide spectrum of host plants, ranging from eudicots to monocots. Interestingly, fungal root colonization results in disease-producing phenotypes only in certain eudicot hosts (Kolander et al., 2012). For instance, F. virguliforme causes sudden death syndrome, one of the most destructive diseases in soybean. Diseased soybean plants exhibit an array of symptoms such as root rot, leaf chlorosis/necrosis, and loss of plant biomass. While several F. virguliforme virulence factors responsible for the disease phenotype have been identified, little is known about how this pathogen evokes symptomatic or asymptomatic responses in different hosts.
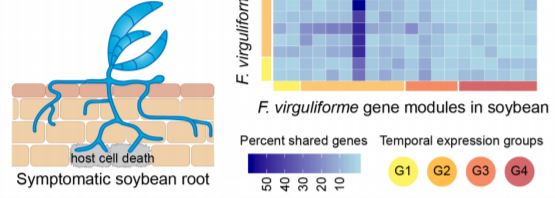
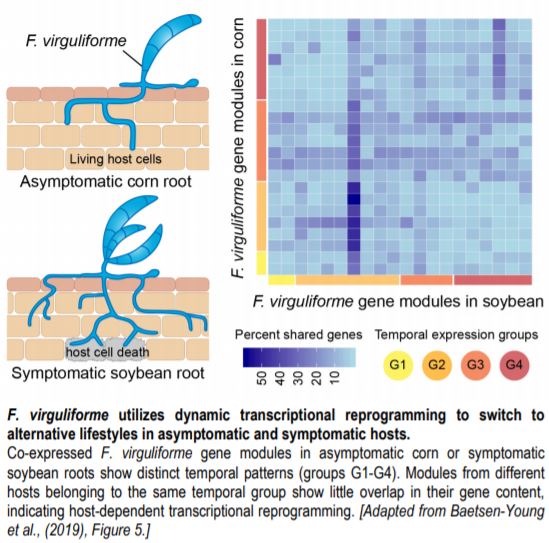
A transcriptome-based study by Baetsen-Young and colleagues (2019) provides a detailed insight into this phenomenon. Instead of looking at a single host-pathogen interaction, the authors compared fungal root colonization and gene expression dynamics in infected soybean (symptomatic) and corn (asymptomatic) hosts. In both of these hosts, germinating F. virguliforme spores can be detected soon after inoculation. Examination of fungal mycelia structures indicated that corn roots undergo a similar, albeit slower infection process to that of soybean roots. However, fungal infection does not trigger host cell death or other negative consequences in corn (see figure), whereas in soybean such symptoms appear as early as 10 days post-inoculation.
 To detect fungal transcripts expressed at the pathogen-host interface, Baetsen-Young et al. (2019) first generated a long read-based high-quality F. virguliforme genome assembly with improved gene annotations. Next, the authors sequenced RNAs from fungal-infected corn or soybean roots (harvested at six different time points during infection), uninfected corn or soybean roots, and in vitro germinated fungal spores without any host. Transcriptome analysis based on a total of 39 RNA-seq libraries revealed that F. virguliforme exhibits both host-specific and inoculation stage-specific gene expression patterns. Compared to the in vitro germinated spores, thousands of fungal genes are differentially expressed in each of the infected hosts. Interestingly, a majority of these fungal genes are highly upregulated (more than four-fold) in both hosts around 7-14 d post-inoculation. Infected corn and soybean roots share little overlap in terms of which fungal genes are upregulated at any given time point. However, in general, fungal genes important for growth and host colonization are induced in both hosts. In soybean roots, fungal genes involved in protein dephosphorylation and small molecule biosynthesis are also upregulated, suggesting that these may be important for symptomatic fungal infection.
To detect fungal transcripts expressed at the pathogen-host interface, Baetsen-Young et al. (2019) first generated a long read-based high-quality F. virguliforme genome assembly with improved gene annotations. Next, the authors sequenced RNAs from fungal-infected corn or soybean roots (harvested at six different time points during infection), uninfected corn or soybean roots, and in vitro germinated fungal spores without any host. Transcriptome analysis based on a total of 39 RNA-seq libraries revealed that F. virguliforme exhibits both host-specific and inoculation stage-specific gene expression patterns. Compared to the in vitro germinated spores, thousands of fungal genes are differentially expressed in each of the infected hosts. Interestingly, a majority of these fungal genes are highly upregulated (more than four-fold) in both hosts around 7-14 d post-inoculation. Infected corn and soybean roots share little overlap in terms of which fungal genes are upregulated at any given time point. However, in general, fungal genes important for growth and host colonization are induced in both hosts. In soybean roots, fungal genes involved in protein dephosphorylation and small molecule biosynthesis are also upregulated, suggesting that these may be important for symptomatic fungal infection.
Global coexpression network analyses identified multiple fungal gene modules associated with F. virguliforme–corn or F. virguliforme–soybean interactions. In both hosts, these modules can be assigned to one of four major groups based on distinct temporal patterns. However, modules belonging to the same temporal group vary greatly in their gene content between the two hosts (see figure). In the case of soybean, gene ontology terms enriched in the temporal groups 1 and 2 correlate to the faster fungal colonization phenotype of this host. Additionally, the temporal expression patterns of these groups reflect fungal transcriptome reprogramming to facilitate the transition from initial manipulation of soybean host defense to subsequent necrotrophic mode for fungal proliferation. At the same time, fungal gene modules upregulated in corn were enriched for processes involved in host-nutrient acquisition. To assess whether host physiology shapes F. virguliforme transcriptome, Baetsen-Young et al. (2019) directly compared fungal gene expressions between corn and soybean for specific time points. This further confirmed distinct changes in fungal transcriptome that are linked to the manifestation of symptomatic or asymptomatic phenotypes. For example, F. virguliforme expresses a greater diversity of necrosis-inducing effectors and pectin-degrading enzymes at a much higher level in infected soybean roots than in corn roots. Soybean has a higher pectin content than corn, so the fungi might produce relatively more pectin lyase enzymes to facilitate host cell breakdown in soybean.
Overall, the findings of Baetsen-Young et al. (2019) show that rather than expressing unique transcripts in different hosts, F. virguliforme extensively reprograms its transcriptome to adopt a necrotrophic or biotrophic lifestyle in soybean or corn, respectively. This transcriptional plasticity might be a common strategy for generalist pathogens to ensure survival on a wide range of morphologically and biochemically diverse hosts in the absence of a compatible host nearby. Questions that remain to be answered include how F. virguliforme accomplishes this temporal regulation, and whether host gene expression is also differentially manipulated by F. virguliforme during asymptomatic vs. symptomatic colonization.
Saima Shahid
Donald Danforth Plant Science Center
Saint Louis, Missouri
ORCID: 0000-0001-9385-0925
REFERENCES
Kolander, T.M., Bienapfl, J.E., Kurle, J.E., and Malvick, D.K. (2012). Symptomatic and asymptomatic host range of Fusarium virguliforme, the causal agent of soybean sudden death syndrome. Plant Dis. 96: 1148-1153.
Baetsen-Young A., Wai C.M., VanBuren R., and Day B. (2019). Fusarium virguliforme transcriptional plasticity revealed by diverged host colonization. Plant Cell. https://doi.org/10.1105/tpc.19.00697