Plant Science Research Weekly: January 31
Review: Characteristics, drivers and feedbacks of global greening
 Whenever we talk about global trends there are of course local variations. Here, Piao et al. discuss recent trends of global greening, which is occurring at arctic as well as temperate and tropical regions. Overall, this greening is attributed to increasing atmospheric CO2, which promotes plant growth, but artic greening is also a consequence of rising temperatures. Human activities including tree planting (afforestation) and nitrogen deposition also contribute to greening, with varying degrees across the globe. The article also discusses the effects of increased greening on water cycles and global temperatures. (Summary by Mary Williams) Nat. Rev. Earth Environ. 10.1038/s43017-019-0001-x
Whenever we talk about global trends there are of course local variations. Here, Piao et al. discuss recent trends of global greening, which is occurring at arctic as well as temperate and tropical regions. Overall, this greening is attributed to increasing atmospheric CO2, which promotes plant growth, but artic greening is also a consequence of rising temperatures. Human activities including tree planting (afforestation) and nitrogen deposition also contribute to greening, with varying degrees across the globe. The article also discusses the effects of increased greening on water cycles and global temperatures. (Summary by Mary Williams) Nat. Rev. Earth Environ. 10.1038/s43017-019-0001-x
AuxSen: a biosensor for direct visualization of auxin
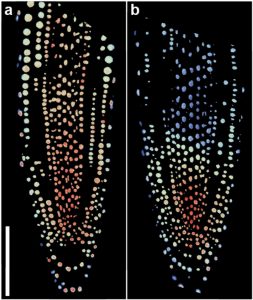 Auxins participate in nearly every aspect of plants’ life cycle, but the information about the actual distribution of this hormone is scarce. The location of the hormone has been inferred using reporters based on auxin-transcriptional responses or auxin-dependent degradation signals. For a direct visualization, Herud-Sikimic and colleagues developed AuxSen: a genetically encoded, reversible biosensor for in vivo imaging of auxin distribution. The authors exploited the structural similarities between indole-3-acetic acid (IAA) and tryptophan. They started from the bacterial tryptophan repressor (TrpR), a tryptophan-binding protein. To increase the specificity of IAA binding and abolishing TRP binding, they generated about 2,000 TrpR variants, and they tested fluorophore and linker combinations to improve the FRET readout. In vitro, the FRET ratio of AuxSen changed upon treatment with IAA. For in vivo analyses, the authors expressed the sensor in protoplasts; the FRET ratio increased proportionally to auxin levels. Additionally, the authors measured the perturbation of auxin caused by inhibition of auxin transport (treatments with NPA and BFA). Two important features of this sensor are: AuxSen expression did not interfere with auxin signaling and its auxin-binding reversibility. With this, mapping of auxin dynamics and concentrations at high cellular resolution is now possible. (Summary by Humberto Herrera-Ubaldo) bioRxiv 10.1101/2020.01.19.911735
Auxins participate in nearly every aspect of plants’ life cycle, but the information about the actual distribution of this hormone is scarce. The location of the hormone has been inferred using reporters based on auxin-transcriptional responses or auxin-dependent degradation signals. For a direct visualization, Herud-Sikimic and colleagues developed AuxSen: a genetically encoded, reversible biosensor for in vivo imaging of auxin distribution. The authors exploited the structural similarities between indole-3-acetic acid (IAA) and tryptophan. They started from the bacterial tryptophan repressor (TrpR), a tryptophan-binding protein. To increase the specificity of IAA binding and abolishing TRP binding, they generated about 2,000 TrpR variants, and they tested fluorophore and linker combinations to improve the FRET readout. In vitro, the FRET ratio of AuxSen changed upon treatment with IAA. For in vivo analyses, the authors expressed the sensor in protoplasts; the FRET ratio increased proportionally to auxin levels. Additionally, the authors measured the perturbation of auxin caused by inhibition of auxin transport (treatments with NPA and BFA). Two important features of this sensor are: AuxSen expression did not interfere with auxin signaling and its auxin-binding reversibility. With this, mapping of auxin dynamics and concentrations at high cellular resolution is now possible. (Summary by Humberto Herrera-Ubaldo) bioRxiv 10.1101/2020.01.19.911735
Putative cis-regulatory elements predict iron deficiency responses in Arabidopsis roots
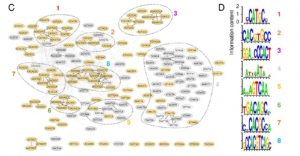 Iron (Fe) is an important micronutrient needed by plants for survival. Plants have evolved a range of morphological, physiological and molecular responses to Fe availability, including the transcriptional regulation of over one thousand genes in response to Fe deficiency (-Fe) in Arabidopsis. However, the cis-regulatory elements (CREs) and their combinations that regulate the adaptive response to -Fe remain largely unknown. Schwarz and colleagues used computational models to identify putative CREs during -Fe. Arabidopsis root transcriptomics and co-expression data were used to identify more than one hundred potential CREs that are indicative of -Fe induced gene expression. Multiple gene expression data sets were used to define iron deficiency co-expression clusters, and a model for their regulation was developed using a machine learning technique. Common CREs across co-expression clusters were identified and compared to known transcription factor binding motifs. Interestingly, the authors found evidence supporting a role in Arabidopsis for the IDE1 element known to have a role in grasses, which use a different strategy for iron uptake. Taken together, Schwarz et al. provided a comprehensive source of cis-regulatory information predictive of iron deficiency responsive genes in Arabidopsis roots. (Summary by Adenike Oyekunle) Plant Physiol. 10.1104/pp.19.00760
Iron (Fe) is an important micronutrient needed by plants for survival. Plants have evolved a range of morphological, physiological and molecular responses to Fe availability, including the transcriptional regulation of over one thousand genes in response to Fe deficiency (-Fe) in Arabidopsis. However, the cis-regulatory elements (CREs) and their combinations that regulate the adaptive response to -Fe remain largely unknown. Schwarz and colleagues used computational models to identify putative CREs during -Fe. Arabidopsis root transcriptomics and co-expression data were used to identify more than one hundred potential CREs that are indicative of -Fe induced gene expression. Multiple gene expression data sets were used to define iron deficiency co-expression clusters, and a model for their regulation was developed using a machine learning technique. Common CREs across co-expression clusters were identified and compared to known transcription factor binding motifs. Interestingly, the authors found evidence supporting a role in Arabidopsis for the IDE1 element known to have a role in grasses, which use a different strategy for iron uptake. Taken together, Schwarz et al. provided a comprehensive source of cis-regulatory information predictive of iron deficiency responsive genes in Arabidopsis roots. (Summary by Adenike Oyekunle) Plant Physiol. 10.1104/pp.19.00760
GPA5 encodes a Rab5a effector required for post-Golgi trafficking of rice storage proteins ($)
 Storage proteins secreted into vacuoles during seed development serve as precursors for germination and plant growth. In this paper, Ren et al. identified a regulator, GLUTELIN PRECURSOR ACCUMULATION 5 (GPA5) that is involved in protein trafficking into the vacuoles during seed development. The loss of function mutant gpa5 was identified through a mutant screen for defects in the vacuolar storage protein transport pathway resulting in the accumulation of glutelin precursor in the apoplast. GPA5 encodes for a phox homology domain-containing protein and is primarily expressed in the prevacuolar compartment/dense vesicles that is necessary for protein trafficking from Golgi to storage vacuoles during seed development. Further, the authors have identified that GPA5 interacts with other regulators of the protein sorting pathway, Rab5a and VPS9a and expression of GPA5 is altered in rab5a and vps9a mutants. Also, GPA5 interacts with a class C core vacuole/endosome tethering (CORVET) complex and VAMP727-containing R-soluble NSF attachment protein receptor (SNARE) complex. Thus, the authors identified a key regulator, GPA5 involved in sorting of storage proteins into vacuoles of developing seeds. (Summary by Suresh Damodaran) Plant Cell 10.1105/tpc.19.00863
Storage proteins secreted into vacuoles during seed development serve as precursors for germination and plant growth. In this paper, Ren et al. identified a regulator, GLUTELIN PRECURSOR ACCUMULATION 5 (GPA5) that is involved in protein trafficking into the vacuoles during seed development. The loss of function mutant gpa5 was identified through a mutant screen for defects in the vacuolar storage protein transport pathway resulting in the accumulation of glutelin precursor in the apoplast. GPA5 encodes for a phox homology domain-containing protein and is primarily expressed in the prevacuolar compartment/dense vesicles that is necessary for protein trafficking from Golgi to storage vacuoles during seed development. Further, the authors have identified that GPA5 interacts with other regulators of the protein sorting pathway, Rab5a and VPS9a and expression of GPA5 is altered in rab5a and vps9a mutants. Also, GPA5 interacts with a class C core vacuole/endosome tethering (CORVET) complex and VAMP727-containing R-soluble NSF attachment protein receptor (SNARE) complex. Thus, the authors identified a key regulator, GPA5 involved in sorting of storage proteins into vacuoles of developing seeds. (Summary by Suresh Damodaran) Plant Cell 10.1105/tpc.19.00863
Death in cells overlying lateral root primordia facilitates organ growth in Arabidopsis
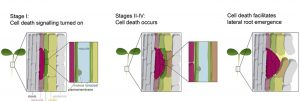 While cell division and cell expansion are known to be determinants of plant organ growth, little is known about the role of cell elimination and death in this process. Cell death indicators such as cell death- and autolysis-associated cysteine proteases have been observed in cells overlying the site of lateral root formation in many plant species, suggesting the involvement of cell death during lateral root emergence. Lateral roots are initiated from pericycle cells that are internal tissues. To emerge, the developing lateral root primordium must pass through the endodermal and cortical cells. It is believed that cell death does not occur during lateral root emergence in Arabidopsis, but rather the overlying cells separate to allow passage. Escamez et al. used a time course gene expression experiment and detected the upregulation of several cell death marker genes in a subset of lateral root primordium overlying cells. Cell-death-detection methods like tonoplast integrity marker, detection of autophagy marker and a viability marker which can distinguish between dead and living cells subsequently confirmed that cell death occurs in a subset of lateral root primordium overlying cells. Perturbation of a central transcriptional regulator of cell death, a NAC transcription factor ORE1 led to a decrease in cell death of lateral root primordium overlying cells and ultimately a delay in lateral root emergence. Interestingly, when cell death was restored by the expression of a mammalian cell death promoting factor or laser ablation, lateral root growth was restored to normal. (Summary by Toluwase Olukayode) Curr. Biol. 10.1016/j.cub.2019.11.078
While cell division and cell expansion are known to be determinants of plant organ growth, little is known about the role of cell elimination and death in this process. Cell death indicators such as cell death- and autolysis-associated cysteine proteases have been observed in cells overlying the site of lateral root formation in many plant species, suggesting the involvement of cell death during lateral root emergence. Lateral roots are initiated from pericycle cells that are internal tissues. To emerge, the developing lateral root primordium must pass through the endodermal and cortical cells. It is believed that cell death does not occur during lateral root emergence in Arabidopsis, but rather the overlying cells separate to allow passage. Escamez et al. used a time course gene expression experiment and detected the upregulation of several cell death marker genes in a subset of lateral root primordium overlying cells. Cell-death-detection methods like tonoplast integrity marker, detection of autophagy marker and a viability marker which can distinguish between dead and living cells subsequently confirmed that cell death occurs in a subset of lateral root primordium overlying cells. Perturbation of a central transcriptional regulator of cell death, a NAC transcription factor ORE1 led to a decrease in cell death of lateral root primordium overlying cells and ultimately a delay in lateral root emergence. Interestingly, when cell death was restored by the expression of a mammalian cell death promoting factor or laser ablation, lateral root growth was restored to normal. (Summary by Toluwase Olukayode) Curr. Biol. 10.1016/j.cub.2019.11.078
MicroRNA397b-LACCASE2 module regulates root lignification under water- and phosphate deficiency ($)
 Water and nutrient availability are a major limiting factors for plant productivity. Plant have evolved a series of complex adaptive features that help them to survive in the hostile environment. Upregulation of lignin biosynthetic genes in roots under salt and water stress have been reported in some plant species, and in Arabidopsis roots during phosphate starvation, yet our knowledge of how lignification is regulated in roots in response to abiotic stress remain elusive. LACCASEs are multi-copper oxidases that catalyze the formation or degradation of various polymers. Khandal et al. identified two LACCASE genes LAC2 and LAC4 whose expression consistently decreases in response to mannitol treated Col-0 arabidopsis seedlings. T-DNA insertion lines of these gene were used to investigate their role in primary root lignification. DAB staining and quantification in mutants and complementation lines showed that LAC2 is a negative regulator of lignification in roots. They also showed that LAC2 expression and root lignification is regulated at the post-transcriptional level by the microRNA miR397b. The authors also showed that LAC2-dependent lignification is not specific to water deficit stress as LAC2 and LAC4 expression was also significantly downregulated in phosphate stress treatment. Genetic testing involving T-DNA mutants, overexpression lines, ad short tandem target mimic were subsequently used to confirm the direct role of miR397b-LAC2 module in drought and phosphate mediated root growth inhibition. (Summary by Toluwase Olukayode) Plant Physiol. 10.1104/pp.19.00921
Water and nutrient availability are a major limiting factors for plant productivity. Plant have evolved a series of complex adaptive features that help them to survive in the hostile environment. Upregulation of lignin biosynthetic genes in roots under salt and water stress have been reported in some plant species, and in Arabidopsis roots during phosphate starvation, yet our knowledge of how lignification is regulated in roots in response to abiotic stress remain elusive. LACCASEs are multi-copper oxidases that catalyze the formation or degradation of various polymers. Khandal et al. identified two LACCASE genes LAC2 and LAC4 whose expression consistently decreases in response to mannitol treated Col-0 arabidopsis seedlings. T-DNA insertion lines of these gene were used to investigate their role in primary root lignification. DAB staining and quantification in mutants and complementation lines showed that LAC2 is a negative regulator of lignification in roots. They also showed that LAC2 expression and root lignification is regulated at the post-transcriptional level by the microRNA miR397b. The authors also showed that LAC2-dependent lignification is not specific to water deficit stress as LAC2 and LAC4 expression was also significantly downregulated in phosphate stress treatment. Genetic testing involving T-DNA mutants, overexpression lines, ad short tandem target mimic were subsequently used to confirm the direct role of miR397b-LAC2 module in drought and phosphate mediated root growth inhibition. (Summary by Toluwase Olukayode) Plant Physiol. 10.1104/pp.19.00921
Three previously characterized resistances to yellow rust are encoded by a single locus Wtk1
 Yellow rust is a fungal disease of wheat. Three mutants with enhanced resistance were identified in wild emmer wheat from different locations were previously mapped to the same region of chromosome 1B. Given that each locus showed a different degree of resistance, it was presumed that they were non-allelic. Klymiuk et al. have now shown that they are all encoded by a single locus, which encodes a protein kinase, Wheat Tandem Kinase 1 (WTK1). Additional studies suggest that the variability in phenotype associated with the different alleles is a consequence of the genetic background in which they were identified rather than being allele-specific. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/eraa020
Yellow rust is a fungal disease of wheat. Three mutants with enhanced resistance were identified in wild emmer wheat from different locations were previously mapped to the same region of chromosome 1B. Given that each locus showed a different degree of resistance, it was presumed that they were non-allelic. Klymiuk et al. have now shown that they are all encoded by a single locus, which encodes a protein kinase, Wheat Tandem Kinase 1 (WTK1). Additional studies suggest that the variability in phenotype associated with the different alleles is a consequence of the genetic background in which they were identified rather than being allele-specific. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/eraa020
Perception of Agrobacterium tumefaciens flagellin by FLS2XL confers resistance to crown gall disease
 FLS2 is a well-characterized cell-surface receptor that recognizes a short epitope found on most bacterial flagellin proteins. The plant pathogen Agrobacterium tumefaciens, causative agent of crown gall disease, deviates strongly at this epitope region, and so is generally not recognized by FLS2 receptors, facilitating its pathogenicity. Fürst et al. set out to try to identify novel FLS2 genes that might recognize this epitope from Agrobacterium tumefaciens. They found a wild grape species, Vitis riparia, that is resistant to the pathogen and has both a standard and a non-standard FLS2 gene. The non-standard variant recognizes both standard epitope and the Agrobacterium tumefaciens epitope, and was named FLS2XL (for extended ligand). Introducing FLS2XL into tobacco plants conferred resistance to crown gall disease. This new finding can provide a new strategy for resistance in plants susceptible to crown gall including fruit trees, raspberry bushes, and grapevines. (Summary by Mary Williams) Nature Plants 10.1038/s41477-019-0578-6
FLS2 is a well-characterized cell-surface receptor that recognizes a short epitope found on most bacterial flagellin proteins. The plant pathogen Agrobacterium tumefaciens, causative agent of crown gall disease, deviates strongly at this epitope region, and so is generally not recognized by FLS2 receptors, facilitating its pathogenicity. Fürst et al. set out to try to identify novel FLS2 genes that might recognize this epitope from Agrobacterium tumefaciens. They found a wild grape species, Vitis riparia, that is resistant to the pathogen and has both a standard and a non-standard FLS2 gene. The non-standard variant recognizes both standard epitope and the Agrobacterium tumefaciens epitope, and was named FLS2XL (for extended ligand). Introducing FLS2XL into tobacco plants conferred resistance to crown gall disease. This new finding can provide a new strategy for resistance in plants susceptible to crown gall including fruit trees, raspberry bushes, and grapevines. (Summary by Mary Williams) Nature Plants 10.1038/s41477-019-0578-6