Timing is Everything: Tandem Fluorescent Timers Expand Our Understanding of Protein Longevity
Signaling within and between cells is fast and dynamic. Such signals can involve the transport of charged ions that change membrane polarity or intercellular gradients, protein de/phosphorylation, protein interactions, and phytohormones, among others. The change in protein distribution in response to these signals is hard to measure, particularly in real time, which limits our understanding of how molecular signals contribute to plant development. Pulse-chase techniques have been used in the past to characterize protein longevity, but these techniques often cannot be conducted in living cells or require chemicals that can have secondary effects on cell function (Claydon and Beynon, 2012; Holman et al., 2016; Nelson et al., 2014; Worley et al., 2000). In this issue of Plant Physiology, Zhang et al. (2019) describe and illustrate how tandem fluorescent protein timers (tFTs) can help distinguish between protein synthesis and degradation in real time, providing a powerful addition to our plant cell biology toolbox.
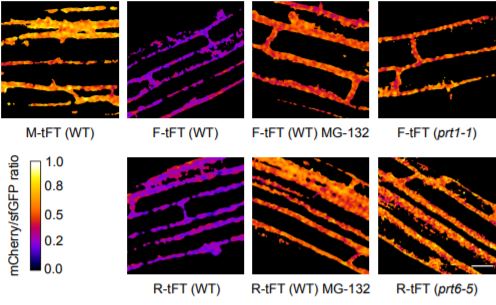
Unlike photo-switchable fluorescent proteins, which change from one emission spectrum to another upon specific excitation, fluorescent timers change color depending on the age of the protein. Recent advances have adapted the use of both fast- and slow-maturing fluorescent proteins so that protein age can be quantified efficiently during the experimental process (Khmelinskii and Knop, 2014; Khmelinskii et al., 2016). In this case, a protein fusion incorporating slow-folding mCherry and fast superfolding GFP (sfGFP) was used to report relative protein lifetimes; as the fusion protein is synthesized, the sfGFP signal is observed first and the mCherry signal is detected as the protein matures. Zhang et al. show that this tFT can be used to measure protein lifetimes both transiently and in stable plant lines to accommodate differential cell types and stimulus responses in vivo. The authors show that their tFT construct could faithfully report protein localization and could indicate endogenous proteasome-dependent protein degradation based on the N-terminal rule, in which N-terminal amino acids are recognized by E3 ligases and target proteins for proteasome degradation (Bachmair et al., 1986; Gibbs et al., 2014). This initial work by the authors supports the stability and usefulness of the tFT because it means the timer itself does not interfere with natural protein degradation pathways and can be applied to different protein kinetic investigations. Moreover, using the Gateway cloning system, this tFT can be applied to a variety of proteins of interest in the future.
Where the rubber really hits the road is the application of the tFT to auxin/IAA proteins to show how the timers can be used to evaluate how proteins with varying lifetimes respond to transient and dynamic early auxin signals (Abel 1994). Zhang et al. show that the tFT-IAA constructs transiently expressed in Nicotiana benthamiana and Arabidopsis thaliana responded to exogenous auxin treatments. In stable transgenic lines, the lifetimes of tFT-IAA constructs also responded to auxin concentration and gravity, indicating that they could report protein stability in real time under changing environmental conditions. The authors do a great job at showing how tFT–protein fusions can be tailored to visualize protein synthesis, localization, responses to stimuli, and degradation with fidelity and little-to-no side effects. This is not trivial: in vitro or static/inhibitory methods of protein turnover measurement do not allow this kind of observation of endogenous protein kinetics. The tFT system definitely improves our understanding and visualization of protein dynamics on a more natural level. Moreover, the work with the Aux/IAA proteins provides some interesting potential applications not only of the tFT, but of these proteins and their degrons as new auxin sensors.
This report opens several new doors to how tFTs can be used in plants with a variety of applications for investigating protein localization and turnover. Their elegant work with the auxin signaling proteins and with mutants should light a creative fire in researchers looking to develop new mutant screens, high throughput or transient expression experiments, and studies of the dynamics of protein lifetimes.
References
Bachmair, A., Finley, D., and Varshavsky, A. (1986). In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186.
Claydon, A.J., and Beynon, R. (2012). Proteome dynamics: revisiting turnover with a global perspective. Mol. Cell. Proteomics MCP 11, 1551–1565.
Gibbs, D.J., Bacardit, J., Bachmair, A., and Holdsworth, M.J. (2014). The eukaryotic N-end rule pathway: conserved mechanisms and diverse functions. Trends Cell Biol. 24, 603–611.
Holman, S.W., Hammond, D.E., Simpson, D.M., Waters, J., Hurst, J.L., and Beynon, R.J. (2016). Protein turnover measurement using selected reaction monitoring-mass spectrometry (SRM-MS). Philos. Transact. A Math. Phys. Eng. Sci. 374.
Khmelinskii, A., and Knop, M. (2014). Analysis of protein dynamics with tandem fluorescent protein timers. Methods Mol. Biol. Clifton NJ 1174, 195–210.
Khmelinskii, A., Meurer, M., Ho, C.-T., Besenbeck, B., Füller, J., Lemberg, M.K., Bukau, B., Mogk, A., and Knop, M. (2016). Incomplete proteasomal degradation of green fluorescent proteins in the context of tandem fluorescent protein timers. Mol. Biol. Cell 27, 360–370.
Nelson, C.J., Li, L., and Millar, A.H. (2014). Quantitative analysis of protein turnover in plants. Proteomics 14, 579–592.
Worley, C.K., Zenser, N., Ramos, J., Rouse, D., Leyser, O., Theologis, A., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. Cell Mol. Biol. 21, 553–562.
Zhang, H., Lister, E., Gannon, L., Leemhuis, W., Rundle, C.A., Theodoulou, F.L., Wirtz, M. (2019). Tandem fluorescent protein timers for non-invasive relative protein lifetime measurement in plants. Plant Phys.