The Way Out: A Transcriptionally Unique Group of Endosperm Cells Implicated in Nutrient Export to the Embryo
Successful development of the maize (Zea mays) kernel requires proper nutrient transport and signaling among its genetically distinct components: the embryo that gives rise to the next generation, the endosperm that nourishes the embryo, and the maternal tissues that surround the embryo and the endosperm (reviewed in Widiez et al., 2017). During its early development, the endosperm differentiates into at least seven cell types with specialized functions, including an embryo-surrounding region (ESR) that presumptively nurtures and defends the embryo (reviewed in Zhan et al., 2017). However, the ESR has degenerated by approximately 14 to 18 days after pollination (DAP) (Widiez et al., 2017), and it has been unclear whether another group of endosperm cells take on the role of the ESR when it is no longer present.
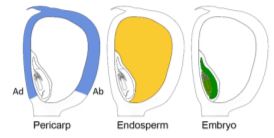
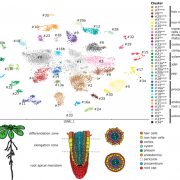
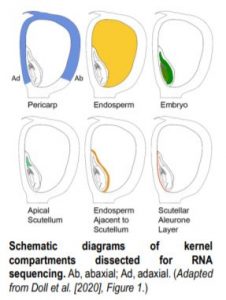 To tackle this question, Doll et al. (2020) performed an RNA-sequencing analysis of two regions of the 13-DAP endosperm that are in direct contact with the adaxial or abaxial side of the embryo, the scutellar aleurone layer (SAL) and the newly named endosperm adjacent to scutellum (EAS), respectively. In addition, the authors also profiled the transcriptomes of the pericarp, whole endosperm, embryo, and apical scutellum at the same developmental stage for comparison (see figure). A principal component analysis of the six kernel compartments revealed that the mRNA populations in the EAS and SAL are both distinct from any other compartment.
To tackle this question, Doll et al. (2020) performed an RNA-sequencing analysis of two regions of the 13-DAP endosperm that are in direct contact with the adaxial or abaxial side of the embryo, the scutellar aleurone layer (SAL) and the newly named endosperm adjacent to scutellum (EAS), respectively. In addition, the authors also profiled the transcriptomes of the pericarp, whole endosperm, embryo, and apical scutellum at the same developmental stage for comparison (see figure). A principal component analysis of the six kernel compartments revealed that the mRNA populations in the EAS and SAL are both distinct from any other compartment.
Differential gene expression analysis followed by gene ontology (GO) enrichment analysis showed that GO terms enriched in gene sets preferentially expressed in the endosperm or embryo were consistent with their developmental stage and known functions. These results indicate that the data accurately reflect the mRNA populations in the profiled compartments. Intriguingly, the set of genes expressed at higher levels in the EAS than in whole endosperm was enriched for the GO term “transmembrane transporter activity.” Similarly, the set of genes preferentially expressed in the SAL also was enriched for GO terms associated with transporter activities. A closer inspection of individual genes associated with the enriched GO terms revealed transporters of amino acids, nucleotides, heavy metals, phosphate, etc. These results suggest strongly that both the EAS and SAL are involved in nutrient transport from endosperm to embryo.
To examine the spatial expression patterns of the genes preferentially expressed in the EAS as compared with whole endosperm, the authors performed mRNA in situ hybridizations for six selected genes using 13-DAP kernels. They showed that, indeed, all six genes were preferentially expressed in the EAS region, supporting the notion that EAS is a transcriptionally unique region. The results also showed that the EAS consists of one to three layers of cells. The authors then examined the temporal dynamics of gene expression in the EAS by carrying out a series of time-course in situ hybridizations on four genes using kernels at five developmental stages spanning 9 to 20 DAP. The data showed that the mRNA levels of these genes consistently peak at 11 to 14 DAP, the stages when the embryo undergoes strong growth, supporting a role for EAS in nourishing the developing embryo. Moreover, the in situ hybridization signals gradually became restricted to the outermost layer of endosperm and eventually disappeared by about 20 DAP, suggesting that the EAS is a spatiotemporally dynamic region. Using terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assays, the authors found that the EAS region undergoes cell death, although it is yet to be determined whether this is triggered by the force from the growing embryo or by the programmed cell death of the endosperm itself.
Finally, the authors investigated whether the direct contact with the embryo scutellum has an impact on the EAS gene expression programs. Using a mutant (emb8522) for which the homozygous seeds contain an aborted embryo, the authors performed in situ hybridizations for four EAS-specific genes using homozygous kernels as well as their wild-type or heterozygous siblings. They found that the presence of a normal embryo represses the ectopic expression of the SWEET15a gene in the SAL, and is necessary for the normal expression of three other genes (PEPB11, Zm00001d017285, and SCL_EAS1) in the EAS.
This study identifies a novel region within the maize endosperm, the endosperm adjacent to scutellum (EAS), that is transcriptionally unique. The previously described cell types in the maize endosperm are characterized by distinct morphological and/or cytological features as well as distinguishable transcriptomes (Zhan et al., 2017). Although no unique cellular feature has been reported for EAS cells, it is tempting to hypothesize that a unique transcriptome might suffice to define them as a new cell type. It is also possible that EAS cells might possess not-yet-characterized cellular characteristics that distinguish them from the other endosperm cell types. Therefore, this study constitutes an important step forward in understanding the interplay between embryo and endosperm as well as cell differentiation within the endosperm.
Junpeng Zhan
Donald Danforth Plant Science Center
St. Louis, Missouri
[email protected]
ORCID ID: 0000-0001-7353-7608
REFERENCES
Doll, N.M., Just, J., Brunaud, V., Caïus, J., Grimault, A., Depège‐Fargeix, N., Esteban, E., Pasha, A., Provart, N.J., Ingram, G.C., Rogowsky, P.M., Widiez, T. (2020). Transcriptomics at maize embryo/endosperm interfaces identify a novel transcriptionally distinct endosperm sub‐domain adjacent to the embryo scutellum. Plant Cell 32: 10.1105/tpc.19.00756.
Widiez, T., Ingram, G.C., and Gutiérrez‐Marcos, J.F. (2017). Embryo‐endosperm‐sporophyte interactions in maize seeds. In Maize Kernel Development, B. Larkins, ed. (Wallingford: CABI), pp. 95–107.
Zhan, J., Dannenhoffer, J.M., and Yadegari, R. (2017). Endosperm development and cell specialization. In Maize Kernel Development, B. Larkins, ed. (Wallingford: CABI), pp. 28–43.