The Trojan Horse Approach to Protein Jockeying
In the decades since Agrobacterium tumefaciens was first used as a vector to deliver genetic material into plants (Zambryski et al., 1983), this powerful tool has provided important insights into the biological functions of countless gene products. However, this approach has its shortcomings; in addition to being time consuming, unpredictable position effects influence the expression and stability of transgenes. Furthermore, as the ability to restrict expression to particular cells is limited, this approach is not ideal for studying small signaling proteins, which carry out their functions in specific cell types at precise stages of development.
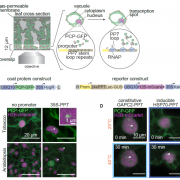
In a Breakthrough Report, van der Linde et al. (2018) introduce an innovative strategy to overcome these hurdles. Exploiting the secretory activity of corn smut fungus (Ustilago maydis), which secretes a range of effector proteins that facilitate infection, the authors developed a system that delivers small proteins of interest into maize (Zea mays) tissue in a highly spatially and temporally restricted manner. In essence, the fungus is engineered to heterologously express a fluorescently-tagged small protein of interest fused to a fungal signal peptide, and expression is driven by a fungal promoter that is active at the desired stage of infection. After infection, the fungus secretes the protein of interest into the plant, where it accumulates in the apoplast of plant cells that come into contact with the fungal hyphae. The researcher then has a window of time in which to study protein function, as normal pathogenic development continues and tumors occur 5–6 days after infection. Because the protein of interest is stealthily smuggled into the host plant, the authors named their approach the Trojan Horse strategy.
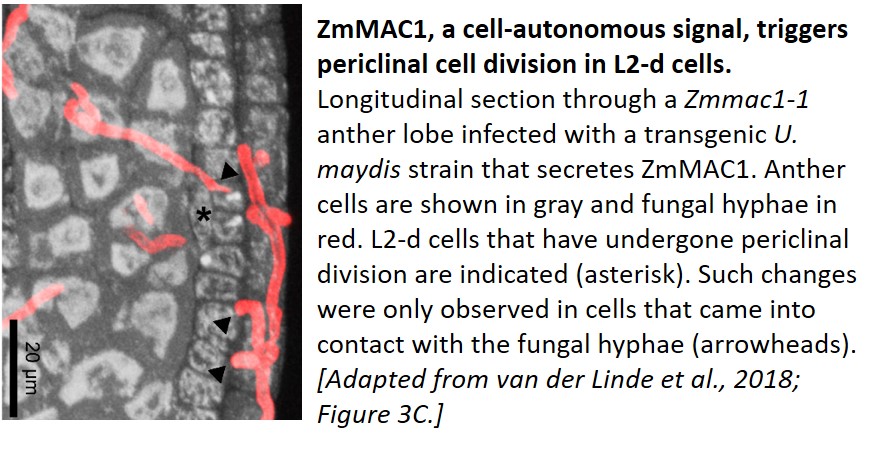
As proof of concept, the authors used their system to introduce the cysteine-rich ZmMAC1 protein, which is thought to initiate somatic niche formation in anthers, into maize plants harboring a mutation in this gene. They infected the plants with a U. maydis strain that generates fluorescently-tagged ZmMAC1 linked to the UmPit2 signal peptide. Because the stage of anther development in maize is faithfully reflected in the length of the anther, delivery of the protein could accurately be timed to a specific developmental stage. Whereas the layer 2-derived (L2-d) cells of the mutant failed to undergo periclinal divisions at a well-documented time point (as described in Wang et al., 2012), those that came into contact with the transgenic fungal hyphae showed a significant increase in periclinal division (see figure). Such an increase was not observed in uninfected neighboring cells, indicating that the protein acts as a cell-autonomous signal. RNA-sequencing and RT-qPCR analysis indicated that anthers infected with the transgenic fungus had somatic niche markers found in wild-type anthers, but not in the mutant, showing that the presence of ZmMAC1 is involved in both cytological and molecular differentiation.
The Trojan Horse strategy provides a valuable tool for rapidly assessing the functions of the dozens of small proteins in maize encoded by unannotated genes. Furthermore, while this study was limited to U. maydis, the authors propose that their approach can be modified to analyze small protein functions in situ in various systems, and list pathogens (and their host plants) that may be suitable for Trojan Horse-mediated small protein delivery.
References
Van der Linde, K., Timofejeva, L., Egger, R.L., Ilau, B., Hammond, R., Teng, C., Meyers, B.C., Doehlemann, G., and Walbot, V. (2018). Pathogen Trojan Horse Delivers Bioactive Host Protein to Alter Maize (Zea mays) Anther Cell Behavior in situ. Plant Cell: 10.1105/tpc.17.00238.
Zambryski, P., Joos, P.H., Genetello, C., Leemans, J., Van Montagu, M., & Schell, J. (1983) Ti plasmid vector for the intriduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J 2: 2143-2150.