The possible danger of divergent non-coding transcription in plants: widespread RNAi
Thieffry et al. give a definitive answer to the question of divergent transcription initiation in Arabidopsis, and discover that if left unchecked, it would lead to rampant RNAi. The Plant Cell (2020) https://doi.org/10.1105/tpc.19.00815
By Albin Sandelin 1,2, Peter Brodersen 1 and Axel Thieffry 1,2, 1 Department of Biology, University of Copenhagen, Ole Maaløes Vej 5, DK2200, Copenhagen N, Denmark; 2 Biotech Research and Innovation Centre, University of Copenhagen, Ole Maaløes Vej 5, DK2200, Copenhagen N, Denmark
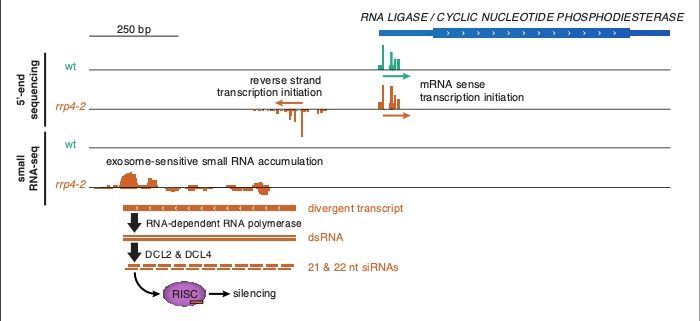
Background: Initiation of transcription is a vital and highly regulated process. Recent research on animal cells has shown that transcription initiation of most genes is accompanied by divergent non-coding transcription on the reverse strand (bidirectional transcription initiation). These reverse strand RNAs are rapidly degraded, making them hard to detect unless relevant degradation pathways are inactivated. Bidirectional transcription has been observed in mammals, insects and fungi, prompting the hypothesis that this is a universal feature of eukaryotes. Surprisingly, plants may be an exception, since some studies found no evidence of bidirectional transcription initiation in plants. However, these studies relied on methods to capture RNAs as they are transcribed and did not use mutants impaired in degradation of aberrant nuclear RNA, including divergent non-coding transcripts.
Question: We investigated the existence of bidirectional transcription initiation in Arabidopsis using mutants in major nuclear RNA degradation pathways. We employed a combination of transcriptomics approaches to map transcription initiation sites at nucleotide resolution, and to uncover the nature of transcripts that accumulate in these mutant backgrounds.
Findings: Our data on Arabidopsis seedlings confirms that plants do not generally employ bidirectional transcription initiation: only around one hundred Arabidopsis genes exhibit this feature. However, our findings strongly indicate that if allowed to accumulate, divergent non-coding transcripts are processed to become small silencing RNAs. This process would depend on RNA-dependent RNA Polymerase, an enzyme found in plants, many fungi, and some invertebrates, but not in mammals or insects. We hypothesize, therefore, that reverse strand transcription initiation in Arabidopsis is selected against, since the resulting RNAs may cause ectopic silencing, potentially including epigenetic silencing via the plant-specific RNA-directed DNA methylation pathway. In addition, our approach revealed a large set of long non-coding anti-sense RNAs whose transcription initiates in the 3’-end regions of protein-coding genes.
Next steps: Our works raises three important questions. 1) Which mechanisms, probably acting at the transcriptional level, eliminate accumulation of divergent non-coding transcripts? 2) Do small interfering RNAs produced from divergent non-coding transcripts really possess detrimental silencing activity, as proposed? 3) Do these non-coding antisense RNAs, or their transcription, play a role in plant gene regulation?
Axel Thieffry et al. (2020). Characterization of Arabidopsis thaliana promoter Bidirectionality and Antisense RNAs by Depletion of Nuclear RNA Decay Pathways. Plant Cell DOI: https://doi.org/10.1105/tpc.19.00815