Discovering Lipid Droplet Proteins—from Seeds to Seedlings
Lynn GL Richardson, Michigan State University, East Lansing, MI 48843
During germination and early seedling establishment, embryos rely on stored energy for growth and cellular maintenance until photoautotrophic growth takes over. Proteins and lipids make up most of the energy-rich stores in the embryo, and in Arabidopsis thaliana, lipids can comprise up to 40% of seed dry weight (Penfield et al., 2005; Baud et al., 2002). Neutral lipids are mainly stored within cytosolic lipid droplets (LDs) as triacylglycerols (TAGs) and sterol esters.
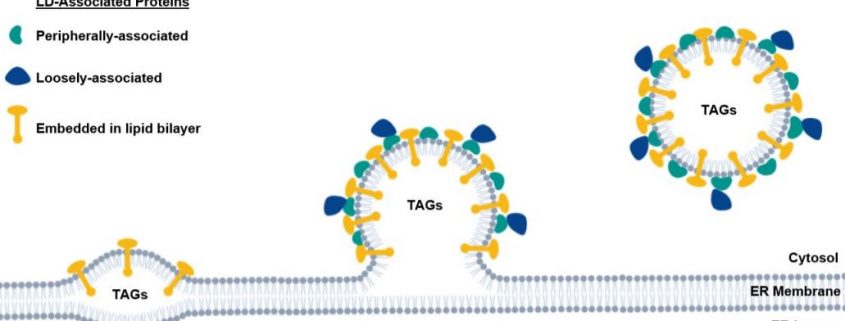
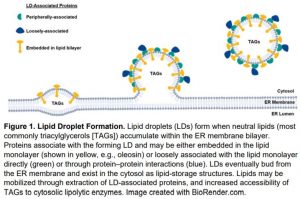
LDs are formed when TAGs and other neutral lipids begin to accumulate within the endoplasmic reticulum (ER) membrane bilayer (Figure 1). The ER membrane leaflet begins to bulge into the cytoplasm as more lipid accumulates, eventually budding off to form the LD (Figure 1) (Pyc et al., 2017; Huang, 2018). The LD is a single phospholipid monolayer surrounded by or embedded with LD-associated proteins (Figure 1). The most abundant LD protein, oleosin, is embedded in the LD membrane and helps maintain lipid droplet structure and regulates lipid mobilization via its extraction and turnover by the ubiquitin-proteasome system (Pyc et al., 2017; Deruyffelaere et al., 2018).
When needed, lipids are released from LDs and mobilized within the plant (Pyc et al., 2017). Lipid mobilization during the early stages of plant development is not well understood, and only a handful of proteins that regulate lipid droplet dynamics have been identified (Pyc et al., 2017; Huang, 2018). It has been suggested that LD proteins act to shield TAGs from lipolytic enzymes; when removed, accessible lipids are extracted and mobilized. Alternatively, LD proteins may interact with cytoplasmic lipases and other regulatory proteins to stimulate lipolysis (Deruyffelaere et al., 2018).
 In this issue of Plant Physiology, Kretzschmar et al., (2019) carried out a quantitative proteomic analysis of LD-associated proteins in Arabidopsis at several early developmental stages: from silique development up until seedling establishment. To identify potential LD proteins, the overall protein abundance profiles were examined over development and in response to desiccation and rehydration. Additionally, a relatively mild technique was developed for enriching LD protein fractions, whereby analysis of an enrichment factor in LD fractions compared to total protein extracts can be used to predict novel, perhaps loosely associated or low-abundance LD proteins (Kretzschmar et al., 2019). Using this method, several novel LD proteins were identified, and their localization to lipid droplets shown using transient expression of fluorescent protein-tagged versions in Nicotiana benthamiana via Agrobacterium infiltration, and in Nicotiana tabacum pollen tubes transformed by biolistic bombardment (Kretzschmar et al., 2019).
In this issue of Plant Physiology, Kretzschmar et al., (2019) carried out a quantitative proteomic analysis of LD-associated proteins in Arabidopsis at several early developmental stages: from silique development up until seedling establishment. To identify potential LD proteins, the overall protein abundance profiles were examined over development and in response to desiccation and rehydration. Additionally, a relatively mild technique was developed for enriching LD protein fractions, whereby analysis of an enrichment factor in LD fractions compared to total protein extracts can be used to predict novel, perhaps loosely associated or low-abundance LD proteins (Kretzschmar et al., 2019). Using this method, several novel LD proteins were identified, and their localization to lipid droplets shown using transient expression of fluorescent protein-tagged versions in Nicotiana benthamiana via Agrobacterium infiltration, and in Nicotiana tabacum pollen tubes transformed by biolistic bombardment (Kretzschmar et al., 2019).
With these combined proteomic/microscopic approaches, LD-association of six proteins was confirmed. These proteins were named based on their functional annotation at The Arabidopsis Information Resource (TAIR), and included a lipase, methyltransferase, hydrolase, dehydrogenase, and an additional LD protein with no functional annotation (Kretzschmar et al., 2019). The functions of these newly discovered LD proteins have yet to be uncovered. The authors noted that the Secretory protein (SEC) 61ɣ subunit of the ER protein translocation machinery (SEC translocase) was also found to be enriched in LD-fractions but did not specifically localize to LDs in the transient expression systems used. The authors note that the ER-localized SEC61ɣ was in close proximity to LDs, which may be indicative of ER-LD junctions (Kretzschmar et al., 2019).
In addition to identifying novel LD-associated proteins, the authors generated a valuable proteomics dataset that will be useful for studying lipid storage and dynamics during seed development and seedling establishment. Many aspects of plant metabolism are now known to be extensively regulated via post-translational mechanisms involving enzymes and/or their associated proteins, rather than through regulation at the transcriptional level (Friso & van Wijk, 2015). As quantitative proteomics approaches continue to improve, these datasets will be instrumental in identifying key regulators of lipid metabolism and other plant metabolic pathways. Combined with previous proteomic analyses (reviewed in Pyc et al., 2017), these dataontribute to our understanding of lipid mobilization and energy partitioning in several economically important species, including oil seed crops (Pyc et al., 2017).
References:
Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160
Deruyffelaere C, Purkrtova Z, Bouchez I, Collet B, Cacas JL, Chardot T, Gallois JL, D’Andrea S (2018) PUX10 is a CDC48A Adaptor Protein that Regulates the Extraction of Ubiquitinated Oleosins from Seed Lipid Droplets in Arabidopsis. Plant Cell 30: 2116-2136
Friso G, van Wijk KJ (2015) Posttranslational Protein Modifications in Plant Metabolism. Plant Phys. 169: 1469-1487
Huang (2018) Plant Lipid Droplets and Their Associated Proteins: Potential for Rapid Advances. Plant Phys. 176: 1894-1918
Penfield S, Graham S, Graham IA (2005) Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem Soc Trans 33:380–383
Pyc M, Cai Y, Greer MS, Yurchenko O, Chapman KD, Dyer JM, Mullen RT (2017) Turning Over a New Leaf in Lipid Droplet Biology. Trends Plant Sci. 22: 596-609
Kretzschamer FK, Doner NM, Krawczyk HE, Scholz P, Schmitt K, Valerius O, Braus GH, Mullen RT, Ischebeck T (2019) Identification of low-abundant lipid droplet proteins in seeds and seedlings. Plant Phys. doi: 10.1104/pp.19.01255