The macroevolutionary history of light signaling ($) (Mol Plant)
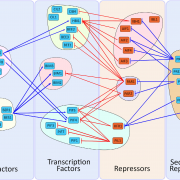
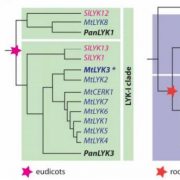
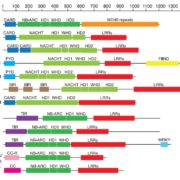
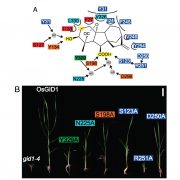
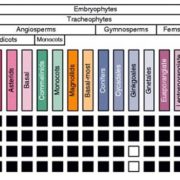
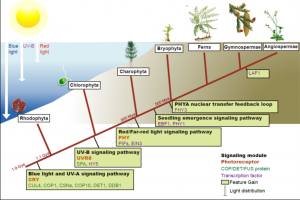 The ability to sense and respond to light is a fundamental feature of photosynthetic organisms like plants. Much has been learned about the molecular genetic mechanisms controlling light perception and downstream signaling processes in evolutionarily young land plant lineages like angiosperms, with comparatively less knowledge in early divergent plants (bryophytes, lycophytes) or their algal predecessors. Han et al. further clarified the evolutionary history of light signaling in land plants and algae by first identifying conserved light signaling genes by surveying the genomes of representative land plant and algal lineages. The authors used this information to reconstruct the evolutionary histories of distinct light signaling modules, the earliest of which appears to be blue-light and UV-A (ultraviolet A) signaling regulated using CRYPTOCHROME (CRY) and the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENESIS1 (COP1) among other genes. Next, UV-B signaling machinery appears first in chlorophytic (marine) algae through the evolution of the UV-B photoreceptor UVR8, the transcriptional regulator HY5 (ELONGATED HYPOCOTYL5), and the general negative regulator of light signaling SUPPRESSOR OF PHY-A (SPA). The red/far-red light signaling pathway appears in charophycean (freshwater) algae by the emergence of PHYTOCHROME receptors (PHY) and PHYTOCHROME INTERACTING FACTORS (PIFs). At around this time in evolutionary history, the authors suggest the acquisition of seedling emergence pathways that allowed buried charophytes/plants to integrate ‘darkness’-associated cues and ethylene signaling to emerge from buried substrate. While functional genetic analyses are required to validate these findings, the comprehensive work of Han et al. illustrates the likely origins and evolutionary history of light signaling modules that were critical for the evolution and expansion of green life on earth. (Summary by Phil Carella) Mol Plant 10.1016/j.molp.2019.04.006
The ability to sense and respond to light is a fundamental feature of photosynthetic organisms like plants. Much has been learned about the molecular genetic mechanisms controlling light perception and downstream signaling processes in evolutionarily young land plant lineages like angiosperms, with comparatively less knowledge in early divergent plants (bryophytes, lycophytes) or their algal predecessors. Han et al. further clarified the evolutionary history of light signaling in land plants and algae by first identifying conserved light signaling genes by surveying the genomes of representative land plant and algal lineages. The authors used this information to reconstruct the evolutionary histories of distinct light signaling modules, the earliest of which appears to be blue-light and UV-A (ultraviolet A) signaling regulated using CRYPTOCHROME (CRY) and the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENESIS1 (COP1) among other genes. Next, UV-B signaling machinery appears first in chlorophytic (marine) algae through the evolution of the UV-B photoreceptor UVR8, the transcriptional regulator HY5 (ELONGATED HYPOCOTYL5), and the general negative regulator of light signaling SUPPRESSOR OF PHY-A (SPA). The red/far-red light signaling pathway appears in charophycean (freshwater) algae by the emergence of PHYTOCHROME receptors (PHY) and PHYTOCHROME INTERACTING FACTORS (PIFs). At around this time in evolutionary history, the authors suggest the acquisition of seedling emergence pathways that allowed buried charophytes/plants to integrate ‘darkness’-associated cues and ethylene signaling to emerge from buried substrate. While functional genetic analyses are required to validate these findings, the comprehensive work of Han et al. illustrates the likely origins and evolutionary history of light signaling modules that were critical for the evolution and expansion of green life on earth. (Summary by Phil Carella) Mol Plant 10.1016/j.molp.2019.04.006