Plant Science Research Weekly: May 17th
Reflections on Classics: Plant Cell‘s 30th anniversary
 “The 1980s were an exciting and revolutionary time for biology, and plant molecular biology in particular,” begins an editorial by Bob Goldberg, Brian Larkins, and Ralph Quatrano, the three Founding Editors of The Plant Cell. They describe why the American Society of Plant Physiologists (ASPP; later, ASPB) felt that the time was right for a new journal that would feature this new discipline. For some of us, 1989 doesn’t seem that long ago; I still remember how our group eagerly gathered around for a first peek at this new journal when I was a PhD student. For others, it may seem incomprehensible that a single cDNA sequence was sufficient for a Plant Cell paper! Looking through the early issues illuminates the strides that have been made in the tools we use to ask “how plants work”. To celebrate the 30th anniversary of The Plant Cell, current and former editors have selected a few of their favorite papers from the journal’s archives and describe how the work has influenced our thinking. The reflections and the papers they highlight can be found here Reflections on Plant Cell Classics. This collection would be useful reading for students! (Summary by Mary Williams). Plant Cell (Editorials 10.1105/tpc.19.00313 and 10.1105/tpc.19.00347).
“The 1980s were an exciting and revolutionary time for biology, and plant molecular biology in particular,” begins an editorial by Bob Goldberg, Brian Larkins, and Ralph Quatrano, the three Founding Editors of The Plant Cell. They describe why the American Society of Plant Physiologists (ASPP; later, ASPB) felt that the time was right for a new journal that would feature this new discipline. For some of us, 1989 doesn’t seem that long ago; I still remember how our group eagerly gathered around for a first peek at this new journal when I was a PhD student. For others, it may seem incomprehensible that a single cDNA sequence was sufficient for a Plant Cell paper! Looking through the early issues illuminates the strides that have been made in the tools we use to ask “how plants work”. To celebrate the 30th anniversary of The Plant Cell, current and former editors have selected a few of their favorite papers from the journal’s archives and describe how the work has influenced our thinking. The reflections and the papers they highlight can be found here Reflections on Plant Cell Classics. This collection would be useful reading for students! (Summary by Mary Williams). Plant Cell (Editorials 10.1105/tpc.19.00313 and 10.1105/tpc.19.00347).
Review. A fruitful journey: Pollen tube navigation from germination to fertilization ($)
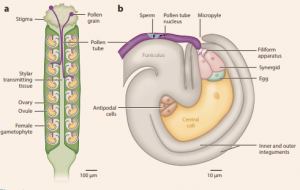 When pollen is deposited on a flower’s stigma, pollen tubes penetrate the stigma and elongate towards ovules where two non-motile sperm will double fertilize an egg and a central cell, which requires cooperation between many cell types. Johnson et al. review our current understanding of these processes. Ca2+ circuits mediate the signaling events that make pollen tube growth possible, through involvement in cytoskeletal dynamics and regulating genes that encode proteins associated with Ca2+ signaling. LURE proteins attract pollen tube growth towards ovules and are species-specific to ensure the correct pollen tube reaches the ovule. Sperm is released when pollen tubes reach the filiform apparatus, a membrane-rich area within the ovule where signaling peptides accumulate along with other small molecules that facilitate pollen tube reception. Sperm fuses with the female gametes by way of HAP2–GCS1, a protein similar in function to the protein that allows some enveloped viruses to enter their hosts. Ca2+ signaling is highly susceptible to temperature stress, making the study of pollen tube growth especially pertinent in the face of increasing temperatures and extreme weather events as byproducts of climate change. (Summary by Rebecca Hayes). Annu. Rev. Plant Biol. 10.1146/annurev-arplant-050718-100133
When pollen is deposited on a flower’s stigma, pollen tubes penetrate the stigma and elongate towards ovules where two non-motile sperm will double fertilize an egg and a central cell, which requires cooperation between many cell types. Johnson et al. review our current understanding of these processes. Ca2+ circuits mediate the signaling events that make pollen tube growth possible, through involvement in cytoskeletal dynamics and regulating genes that encode proteins associated with Ca2+ signaling. LURE proteins attract pollen tube growth towards ovules and are species-specific to ensure the correct pollen tube reaches the ovule. Sperm is released when pollen tubes reach the filiform apparatus, a membrane-rich area within the ovule where signaling peptides accumulate along with other small molecules that facilitate pollen tube reception. Sperm fuses with the female gametes by way of HAP2–GCS1, a protein similar in function to the protein that allows some enveloped viruses to enter their hosts. Ca2+ signaling is highly susceptible to temperature stress, making the study of pollen tube growth especially pertinent in the face of increasing temperatures and extreme weather events as byproducts of climate change. (Summary by Rebecca Hayes). Annu. Rev. Plant Biol. 10.1146/annurev-arplant-050718-100133
Exome sequencing highlights the role of wild-relative introgression in the wheat genome, and Tracing the ancestry of modern bread wheats
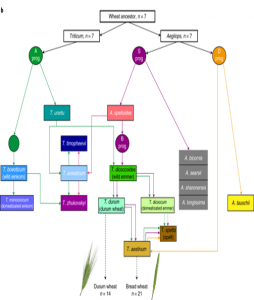 Wheat is of course a hugely important food for humans, and has been selectively bred across the globe for millennia. Modern bread wheats are hexaploid and contain three distinct subgenomes (AABBDD). As with other crops, there is a need to understand wheat’s ancestry and explore the greater genetic diversity of its extended family as sources of beneficial genes. A pair of back-to-back papers in Nature Genetics focus on wheat’s ancestry. He et al. sequenced exomes from 890 accessions of wheat landraces and cultivars, and conclude, “historic gene flow from wild relatives made a substantial contribution to the adaptive diversity of modern bread wheat.” Pont et al. used a similar approach, and conclude that today’s wheat genome has been, “shaped during domestication by human migration, anthropogenic selection and latterly by breeding.” Together, these studies provide, “a rich source of genetic diversity that can be applied to understanding and improving diverse traits, from environmental adaptation to disease resistance and nutrient use efficiency.”(Summary by Mary Williams) Nature Genetics 10.1038/s41588-019-0382-2 and 10.1038/s41588-019-0393-z
Wheat is of course a hugely important food for humans, and has been selectively bred across the globe for millennia. Modern bread wheats are hexaploid and contain three distinct subgenomes (AABBDD). As with other crops, there is a need to understand wheat’s ancestry and explore the greater genetic diversity of its extended family as sources of beneficial genes. A pair of back-to-back papers in Nature Genetics focus on wheat’s ancestry. He et al. sequenced exomes from 890 accessions of wheat landraces and cultivars, and conclude, “historic gene flow from wild relatives made a substantial contribution to the adaptive diversity of modern bread wheat.” Pont et al. used a similar approach, and conclude that today’s wheat genome has been, “shaped during domestication by human migration, anthropogenic selection and latterly by breeding.” Together, these studies provide, “a rich source of genetic diversity that can be applied to understanding and improving diverse traits, from environmental adaptation to disease resistance and nutrient use efficiency.”(Summary by Mary Williams) Nature Genetics 10.1038/s41588-019-0382-2 and 10.1038/s41588-019-0393-z
Identification of transcription factors regulating senescence in wheat through gene regulatory network modelling
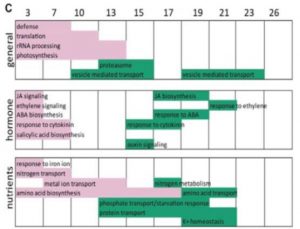 Like other seed crops, wheat yields depend in part on the efficiency with which nutrients stored in leaves are mobilized into the developing seeds. This depends on the several processes from macromolecule breakdown to transport, as well as the timing of leaf senescence. Borrill et al. used RNA analysis and gene regulatory network (GRN) modeling to uncover transcription factors involved in the regulation of leaf senescence. They found nearly 10,000 genes differentially regulated during flag leaf senescence, which fall into early and late temporal waves. Early, down-regulated genes include those involved in photosynthesis and protein translation, whereas late, up-regulated genes include those involved in nutrient mobilization and transport. Among other things, this study shows how far wheat genomic tools have progressed. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00380
Like other seed crops, wheat yields depend in part on the efficiency with which nutrients stored in leaves are mobilized into the developing seeds. This depends on the several processes from macromolecule breakdown to transport, as well as the timing of leaf senescence. Borrill et al. used RNA analysis and gene regulatory network (GRN) modeling to uncover transcription factors involved in the regulation of leaf senescence. They found nearly 10,000 genes differentially regulated during flag leaf senescence, which fall into early and late temporal waves. Early, down-regulated genes include those involved in photosynthesis and protein translation, whereas late, up-regulated genes include those involved in nutrient mobilization and transport. Among other things, this study shows how far wheat genomic tools have progressed. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00380
Origin of angiosperms and the puzzle of the Jurassic gap
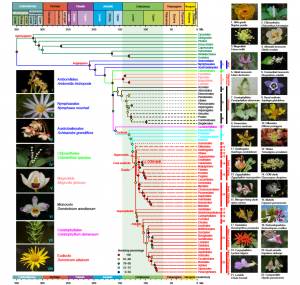 Understanding the origin and evolution of flowering plants is key to explaining the development of major terrestrial ecosystems. The rapid diversification of angiosperms into over 360,000 extant species was famously termed ‘an abominable mystery’ by Charles Darwin. Here, Li et al. reconstruct the angiosperm phylogeny based on 80 genes from 2881 plant plastomes, providing the most comprehensive taxonomic sampling to date. The findings show high similarity to the recent Angiosperm Phylogeny Group (APG IV) report which is based on a consensus of published phylogenetic studies. Dating analysis revealed that angiosperms originated around 209 million years ago (Ma) which is distinct from the oldest fossilised evidence of flowering plants at approximately 135 Ma. The difference between age estimates based on molecular data and the fossil record is coined the Jurassic angiosperm gap. (Summary by Alex Bowles) Nature Plants 10.1038/s41477-019-0421-0
Understanding the origin and evolution of flowering plants is key to explaining the development of major terrestrial ecosystems. The rapid diversification of angiosperms into over 360,000 extant species was famously termed ‘an abominable mystery’ by Charles Darwin. Here, Li et al. reconstruct the angiosperm phylogeny based on 80 genes from 2881 plant plastomes, providing the most comprehensive taxonomic sampling to date. The findings show high similarity to the recent Angiosperm Phylogeny Group (APG IV) report which is based on a consensus of published phylogenetic studies. Dating analysis revealed that angiosperms originated around 209 million years ago (Ma) which is distinct from the oldest fossilised evidence of flowering plants at approximately 135 Ma. The difference between age estimates based on molecular data and the fossil record is coined the Jurassic angiosperm gap. (Summary by Alex Bowles) Nature Plants 10.1038/s41477-019-0421-0
Yam genomics supports West Africa as a major cradle of crop domestication
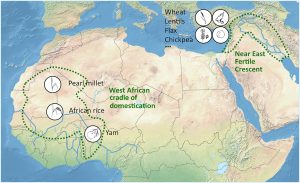 Yams (Dioscorea spp.) were domesticated independently in three continents. African yam (Dioscorea rotundata) is the second most produced crop in Africa, after cassava but ahead of maize, rice and sorghum. Scarcelli et al. use a genomic approach to learn more about its domestication, by sequencing many varieties of the domesticated species as well as its two closest wild relatives. Through this work they identified signatures of selection for several genes contributing to domestication, including genes that would promote larger, starchy tubers. Alongside other studies of African crops, “these results greatly refine our understanding of West African crops domestication history and identify a major cradle of domestication in West Africa, geographically localized around the Niger River, comparable to the Fertile Crescent in the Near East.” (Summary by Mary Williams) Sci Advances 10.1126/sciadv.aaw1947
Yams (Dioscorea spp.) were domesticated independently in three continents. African yam (Dioscorea rotundata) is the second most produced crop in Africa, after cassava but ahead of maize, rice and sorghum. Scarcelli et al. use a genomic approach to learn more about its domestication, by sequencing many varieties of the domesticated species as well as its two closest wild relatives. Through this work they identified signatures of selection for several genes contributing to domestication, including genes that would promote larger, starchy tubers. Alongside other studies of African crops, “these results greatly refine our understanding of West African crops domestication history and identify a major cradle of domestication in West Africa, geographically localized around the Niger River, comparable to the Fertile Crescent in the Near East.” (Summary by Mary Williams) Sci Advances 10.1126/sciadv.aaw1947
Development of genome-wide SNP markers for barley via reference- based RNA-Seq Analysis
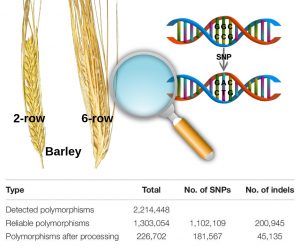 The availability of marker-assisted systems is key for crop improvement, allowing trait selection by identifying consensus polymorphisms. However, for discriminating between related strains, these DNA markers are limited, time-consuming and expensive. Here, Tanaka et al. developed a RNA-Seq-based genotyping pipeline for genome-wide polymorphism detection for barley. The authors compared the variations among 108 Japanese strains of two and six-row barley, by mapping onto the reference public genome sequence of cultivar Morex (Hordeum vulgare) to test efficiency. Using only the transcript region and their ∼3-Kbp flanking regions, instead of the whole genome, reduces the mapping time by half and the genotyping time by two-thirds. They validated this method by amplicon sequencing (AmpliSeq) and found that AmpliSeq detects a wider variety of polymorphisms than RNA-Seq, yet it requires prior identification of the polymorphisms; thus an optimal approach is the combination of both techniques. This study reveals the efficiency reached by a DNA marker based on RNA-Seq able to compare related strains. As expected, the number of polymorphisms was lower in same than between different row type strains. (Summary by Ana Valladares). Front Plant Sci. 10.3389/fpls.2019.00403
The availability of marker-assisted systems is key for crop improvement, allowing trait selection by identifying consensus polymorphisms. However, for discriminating between related strains, these DNA markers are limited, time-consuming and expensive. Here, Tanaka et al. developed a RNA-Seq-based genotyping pipeline for genome-wide polymorphism detection for barley. The authors compared the variations among 108 Japanese strains of two and six-row barley, by mapping onto the reference public genome sequence of cultivar Morex (Hordeum vulgare) to test efficiency. Using only the transcript region and their ∼3-Kbp flanking regions, instead of the whole genome, reduces the mapping time by half and the genotyping time by two-thirds. They validated this method by amplicon sequencing (AmpliSeq) and found that AmpliSeq detects a wider variety of polymorphisms than RNA-Seq, yet it requires prior identification of the polymorphisms; thus an optimal approach is the combination of both techniques. This study reveals the efficiency reached by a DNA marker based on RNA-Seq able to compare related strains. As expected, the number of polymorphisms was lower in same than between different row type strains. (Summary by Ana Valladares). Front Plant Sci. 10.3389/fpls.2019.00403
Arabidopsis FLL2 promotes liquid–liquid phase separation of polyadenylation complexes ($)
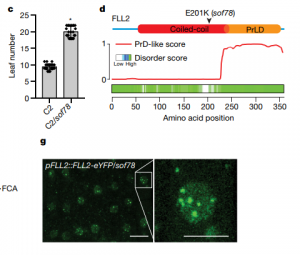 The nucleus of plants cells, like other eukaryotes, is full of non-membranous compartments separated by liquid-liquid phases. These complexes are often called nuclear bodies and concentrate proteins and nucleic acids. Disordered protein domains play a critical in their formation. Here, Fang et al. aimed to study the function of FCA, an Arabidopsis protein required for the alternative 3’-end processing of the long non-coding RNA COOLAIR that regulates the critical flowering regulator Flowering Locus C (FLC). Through a mutagenesis strategy, they searched for factors required for FCA-mediated repression of FLC. They found FLL2, a disordered protein with a coiled-coil domain. This protein colocalizes and interacts with FCA in nuclear bodies through a shared disordered prion-like domain. The mutation of FLL2 reduces the FCA nuclear body localization, and both can promote liquid-liquid phase separation in vitro and possess a liquid-like behavior in vivo. These proteins also are necessary to promote polyadenylation and are associated with the 3′ RNA-processing components. Moreover, their homologs, FLL1 and FLL3, also could carry out similar functions. This paper is a good example of how to approach to the fascinating field of disordered proteins and liquid-liquid phase separations in plants. (Summary by Facundo Romani) Nature 110.1038/s41586-019-1165-8
The nucleus of plants cells, like other eukaryotes, is full of non-membranous compartments separated by liquid-liquid phases. These complexes are often called nuclear bodies and concentrate proteins and nucleic acids. Disordered protein domains play a critical in their formation. Here, Fang et al. aimed to study the function of FCA, an Arabidopsis protein required for the alternative 3’-end processing of the long non-coding RNA COOLAIR that regulates the critical flowering regulator Flowering Locus C (FLC). Through a mutagenesis strategy, they searched for factors required for FCA-mediated repression of FLC. They found FLL2, a disordered protein with a coiled-coil domain. This protein colocalizes and interacts with FCA in nuclear bodies through a shared disordered prion-like domain. The mutation of FLL2 reduces the FCA nuclear body localization, and both can promote liquid-liquid phase separation in vitro and possess a liquid-like behavior in vivo. These proteins also are necessary to promote polyadenylation and are associated with the 3′ RNA-processing components. Moreover, their homologs, FLL1 and FLL3, also could carry out similar functions. This paper is a good example of how to approach to the fascinating field of disordered proteins and liquid-liquid phase separations in plants. (Summary by Facundo Romani) Nature 110.1038/s41586-019-1165-8
Soil salinity limits plant shade avoidance

The agriculture sector is going to face big challenges to feed the 10 billion people that are going to inhabit the planet in the upcoming years with limited arable land. One effective practice to enhance the yield per unit area is to increase crop planting density. However, in dense stands, plants compete with each other for light. They perceive this vegetative shading mainly as a reduction in the ratio of red to far-red light caused by the depletion of photosynthetically active radiation by neighbouring plants. Such a signal serves as a warning of forthcoming competition from neighbours and triggers a number of physiological alterations that are described as the shade avoidance response. Hayes et al. are interested in the connection of this developmental program with other environmental factors. Here they show that even low levels of salt interfere with shade avoidance. In particular they highlight and demonstrate a link between light, ABA-, and BR-signaling pathways. (Summarized by Francesca Resentini) Curr. Biol. 10.1016/j.cub.2019.03.042
The macroevolutionary history of light signaling ($)
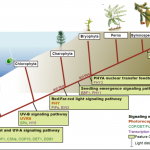
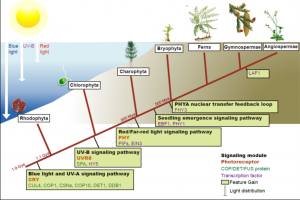 The ability to sense and respond to light is a fundamental feature of photosynthetic organisms like plants. Much has been learned about the molecular genetic mechanisms controlling light perception and downstream signaling processes in evolutionarily young land plant lineages like angiosperms, with comparatively less knowledge in early divergent plants (bryophytes, lycophytes) or their algal predecessors. Han et al. further clarified the evolutionary history of light signaling in land plants and algae. They first identified conserved light signaling genes by surveying the genomes of representative land plant and algal lineages. The authors used this information to reconstruct the evolutionary histories of distinct light signaling modules, the earliest of which appears to be blue-light and UV-A (ultraviolet A) signaling regulated using CRYPTOCHROME (CRY) and the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENESIS1 (COP1) among other genes. Next, UV-B signaling machinery appears first in chlorophytic (marine) algae through the evolution of the UV-B photoreceptor UVR8, the transcriptional regulator HY5 (ELONGATED HYPOCOTYL5), and the general negative regulator of light signaling SUPPRESSOR OF PHY-A (SPA). The red/far-red light signaling pathway appears in charophycean (freshwater) algae by the emergence of PHYTOCHROME receptors (PHY) and PHYTOCHROME INTERACTING FACTORS (PIFs). At around this time in evolutionary history, the authors suggest the acquisition of seedling emergence pathways that allowed buried charophytes/plants to integrate ‘darkness’-associated cues and ethylene signaling to emerge from buried substrate. While functional genetic analyses are required to validate these findings, the comprehensive work of Han et al. illustrates the likely origins and evolutionary history of light signaling modules that were critical for the evolution and expansion of green life on earth. (Summary by Phil Carella) Mol Plant 10.1016/j.molp.2019.04.006
The ability to sense and respond to light is a fundamental feature of photosynthetic organisms like plants. Much has been learned about the molecular genetic mechanisms controlling light perception and downstream signaling processes in evolutionarily young land plant lineages like angiosperms, with comparatively less knowledge in early divergent plants (bryophytes, lycophytes) or their algal predecessors. Han et al. further clarified the evolutionary history of light signaling in land plants and algae. They first identified conserved light signaling genes by surveying the genomes of representative land plant and algal lineages. The authors used this information to reconstruct the evolutionary histories of distinct light signaling modules, the earliest of which appears to be blue-light and UV-A (ultraviolet A) signaling regulated using CRYPTOCHROME (CRY) and the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENESIS1 (COP1) among other genes. Next, UV-B signaling machinery appears first in chlorophytic (marine) algae through the evolution of the UV-B photoreceptor UVR8, the transcriptional regulator HY5 (ELONGATED HYPOCOTYL5), and the general negative regulator of light signaling SUPPRESSOR OF PHY-A (SPA). The red/far-red light signaling pathway appears in charophycean (freshwater) algae by the emergence of PHYTOCHROME receptors (PHY) and PHYTOCHROME INTERACTING FACTORS (PIFs). At around this time in evolutionary history, the authors suggest the acquisition of seedling emergence pathways that allowed buried charophytes/plants to integrate ‘darkness’-associated cues and ethylene signaling to emerge from buried substrate. While functional genetic analyses are required to validate these findings, the comprehensive work of Han et al. illustrates the likely origins and evolutionary history of light signaling modules that were critical for the evolution and expansion of green life on earth. (Summary by Phil Carella) Mol Plant 10.1016/j.molp.2019.04.006
Loss-of-function of a tomato receptor-like kinase impairs male fertility and induces parthenocarpic fruit set
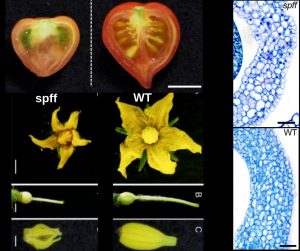 The flower-to-fruit transition (fruit set) and parthenocarpy, the pollination-independent development of seedless fruits, are controlled by complex hormone networks. Takei et al. identified a tomato gamma-ray induced mutant named small parthenocarpic fruit and flower (spff) with small fruits and flowers, male sterility, delayed flowering, and increased productivity (fruits per plant). Using genome-wide single nucleotide polymorphism arrays, mapping-by-sequencing and further validation by Knock-Down mutants using RNA interference (RNAi), the authors identified a monogenic recessive mutation in the Solyc04g077010 gene, which encodes a RLK protein (receptor-like kinase). The mutation associated is a 2bp deletion, which causes a frame-shift generating a truncated, loss-of-function RLK. Ovarian transcriptome analysis of spff lines revealed increased GA20ox1 levels, which is one of the key factors involved in gibberellin (GA) biosynthesis in tomato and which has been related to parthenocarpy. The authors propose that as RLK may repress GA response, the parthenocarpy in spff may be induced by increased GA response. (Summary by Ana Valladares). Front Plant Sci 10.3389/fpls.2019.00403
The flower-to-fruit transition (fruit set) and parthenocarpy, the pollination-independent development of seedless fruits, are controlled by complex hormone networks. Takei et al. identified a tomato gamma-ray induced mutant named small parthenocarpic fruit and flower (spff) with small fruits and flowers, male sterility, delayed flowering, and increased productivity (fruits per plant). Using genome-wide single nucleotide polymorphism arrays, mapping-by-sequencing and further validation by Knock-Down mutants using RNA interference (RNAi), the authors identified a monogenic recessive mutation in the Solyc04g077010 gene, which encodes a RLK protein (receptor-like kinase). The mutation associated is a 2bp deletion, which causes a frame-shift generating a truncated, loss-of-function RLK. Ovarian transcriptome analysis of spff lines revealed increased GA20ox1 levels, which is one of the key factors involved in gibberellin (GA) biosynthesis in tomato and which has been related to parthenocarpy. The authors propose that as RLK may repress GA response, the parthenocarpy in spff may be induced by increased GA response. (Summary by Ana Valladares). Front Plant Sci 10.3389/fpls.2019.00403