The genomic ecosystem of transposable elements in maize (PLOS Genetics)
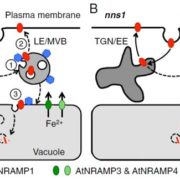
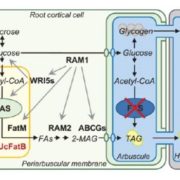
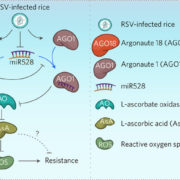
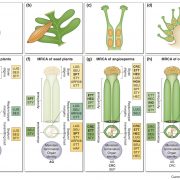
 A new paper looking at transposable elements in maize uses the framework that genomes are similar to ecosystems, in that it is essential to study them comprehensively, from the level of each element to the global structure. Transposable elements (TEs) are dynamic and persistent within plant genomes. Understanding their behavior and physiology can give us a broader perspective of the evolutionary potential of their host genome and species. Stitzer and colleagues explored the Maize B73 reference genome annotation to characterize TE families and their relationships. They annotated and classified more than 27,000 TE families in 13 superfamilies. To build a family survival (age) model, they measured features like TE taxonomy, length, expression, methylation, etc. Many characteristics were shared across families of the same superfamily. Nevertheless, some families had specific contributions to the community. Further, some features correlated to TE age and led the model response. Predictions emphasize studying each family apart since they are adapted to a unique niche. TEs evolve in parallel to maize and are vital components of its evolution and physiology. This work encourages integrated TE ecosystem depiction in our current plant genome analysis. (Summary and sketch by Eddy Mendoza-Galindo @IamSomnya) PLOS Genetics 10.1371/journal.pgen.100976
A new paper looking at transposable elements in maize uses the framework that genomes are similar to ecosystems, in that it is essential to study them comprehensively, from the level of each element to the global structure. Transposable elements (TEs) are dynamic and persistent within plant genomes. Understanding their behavior and physiology can give us a broader perspective of the evolutionary potential of their host genome and species. Stitzer and colleagues explored the Maize B73 reference genome annotation to characterize TE families and their relationships. They annotated and classified more than 27,000 TE families in 13 superfamilies. To build a family survival (age) model, they measured features like TE taxonomy, length, expression, methylation, etc. Many characteristics were shared across families of the same superfamily. Nevertheless, some families had specific contributions to the community. Further, some features correlated to TE age and led the model response. Predictions emphasize studying each family apart since they are adapted to a unique niche. TEs evolve in parallel to maize and are vital components of its evolution and physiology. This work encourages integrated TE ecosystem depiction in our current plant genome analysis. (Summary and sketch by Eddy Mendoza-Galindo @IamSomnya) PLOS Genetics 10.1371/journal.pgen.100976