The genome of broomcorn millet: The fourth millet to be sequenced (Nature Comms)
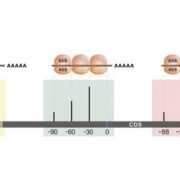
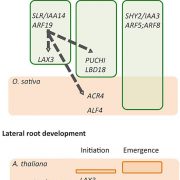
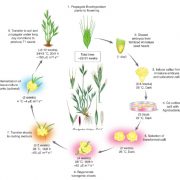
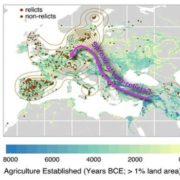
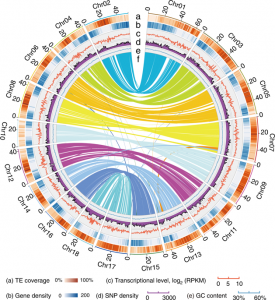 Whole genome sequencing (WGS) unlocks the repertoire of genes encoded within the genome. From a structural genomics aspect, the sequence data enables the development of molecular markers and maps, and identification of novel QTLs/genes/alleles regulating the trait-of-interest. From a functional genomics perspective, the sequence information provides direct access to the genes responsible for the trait-of-interest, which could be used in developing transgenics. WGS has been popular among the ‘known’ crops including rice, wheat, barley, etc. However, a predominant category of plants remains ‘understudied’: these include millets, tubers, legumes and leafy vegetables. Among millets, the genomes of three species have been sequenced, namely, foxtail, pearl and finger millet. Now, broomcorn millet enters the genome club, and of note, broomcorn is the first minor millet to get its genome sequenced. Illumina short-read coupled with Pac-Bio long-read sequencing of this allotetraploid crop (2n = 4X= 36; ~923 Mb) provided the information about 55930 protein-coding genes and 339 microRNA genes present in the genome. In addition, 132 individuals from an F6 population of recombinant inbred lines (RIL) were also sequenced, and a high-density genetic map was constructed using 221787 SNP markers. This iformation will expedite structural and functional genomics studies in broomcorn millet, and also assist crop improvement programs. (Summary by Muthamilarasan Mehanathan). Nature Comms. 10.1038/s41467-019-08409-5
Whole genome sequencing (WGS) unlocks the repertoire of genes encoded within the genome. From a structural genomics aspect, the sequence data enables the development of molecular markers and maps, and identification of novel QTLs/genes/alleles regulating the trait-of-interest. From a functional genomics perspective, the sequence information provides direct access to the genes responsible for the trait-of-interest, which could be used in developing transgenics. WGS has been popular among the ‘known’ crops including rice, wheat, barley, etc. However, a predominant category of plants remains ‘understudied’: these include millets, tubers, legumes and leafy vegetables. Among millets, the genomes of three species have been sequenced, namely, foxtail, pearl and finger millet. Now, broomcorn millet enters the genome club, and of note, broomcorn is the first minor millet to get its genome sequenced. Illumina short-read coupled with Pac-Bio long-read sequencing of this allotetraploid crop (2n = 4X= 36; ~923 Mb) provided the information about 55930 protein-coding genes and 339 microRNA genes present in the genome. In addition, 132 individuals from an F6 population of recombinant inbred lines (RIL) were also sequenced, and a high-density genetic map was constructed using 221787 SNP markers. This iformation will expedite structural and functional genomics studies in broomcorn millet, and also assist crop improvement programs. (Summary by Muthamilarasan Mehanathan). Nature Comms. 10.1038/s41467-019-08409-5