TET-mediated epimutagenesis of the Arabidopsis thaliana methylome
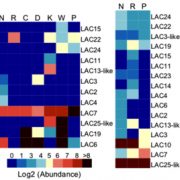
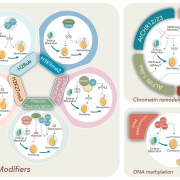
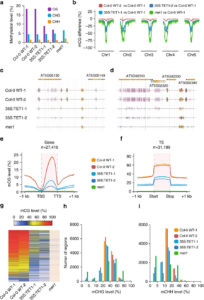 Natural and induced DNA methylation variations are known to alter gene expression changes that may ultimately be important for agronomically important traits. Epigenetically manipulating plant methylomes to create heritable changes can be invaluable in crop improvement programs. Ji et al. describe the expression of a human ten eleven translocation enzyme (TET) in Arabidopsis thaliana to create random demethylation of the genome. The widespread hypomethylation mimics mutants lacking the MET1 DNA methyltransferase, although with less severe phenotypes. hTET1cd expression in Arabidopsis resulted in a global reduction in CG methylation, while its effect was not as severe on CHG and CHH methylation. Individual transgenic lines showed varying levels of reduction in CG methylation, making it a suitable system to select for desired levels of demethylation. Similar to met1, hTET1 plants showed hypomethylation of the seventh intron of INCREASE IN BONSAI METHYLATION 1 (IBM1), a histone H3K9 demethylase, resulting in altered expression and CHG hypermethylation in gene body methylated (gBM) loci. Similarly, they also show loss of methylation at FLOWERING WAGENINGEN (FWA), associated with its increased expression and a delayed flowering phenotype. The authors also show that using the ACTIN 2 promoter (ACT2:sfGFP-hTET1cd) that has activity in all tissue types of juvenile plants substantially delays flowering time in the transgenic plants compared to lines with 35S:hTET1cd. The late flowering phenotype was stably inherited in the progeny irrespective of the presence of transgene. In contrast, in the 35S:hTET1cd, the transgenerational inheritance was minimum after removal of the transgene. TET-mediated engineering of DNA methylation states to create epialleles that are faithfully inherited in subsequent generations in agriculturally important crop species is an interesting area for further investigation. Nature Comms. 10.1038/s41467-018-03289-7
Natural and induced DNA methylation variations are known to alter gene expression changes that may ultimately be important for agronomically important traits. Epigenetically manipulating plant methylomes to create heritable changes can be invaluable in crop improvement programs. Ji et al. describe the expression of a human ten eleven translocation enzyme (TET) in Arabidopsis thaliana to create random demethylation of the genome. The widespread hypomethylation mimics mutants lacking the MET1 DNA methyltransferase, although with less severe phenotypes. hTET1cd expression in Arabidopsis resulted in a global reduction in CG methylation, while its effect was not as severe on CHG and CHH methylation. Individual transgenic lines showed varying levels of reduction in CG methylation, making it a suitable system to select for desired levels of demethylation. Similar to met1, hTET1 plants showed hypomethylation of the seventh intron of INCREASE IN BONSAI METHYLATION 1 (IBM1), a histone H3K9 demethylase, resulting in altered expression and CHG hypermethylation in gene body methylated (gBM) loci. Similarly, they also show loss of methylation at FLOWERING WAGENINGEN (FWA), associated with its increased expression and a delayed flowering phenotype. The authors also show that using the ACTIN 2 promoter (ACT2:sfGFP-hTET1cd) that has activity in all tissue types of juvenile plants substantially delays flowering time in the transgenic plants compared to lines with 35S:hTET1cd. The late flowering phenotype was stably inherited in the progeny irrespective of the presence of transgene. In contrast, in the 35S:hTET1cd, the transgenerational inheritance was minimum after removal of the transgene. TET-mediated engineering of DNA methylation states to create epialleles that are faithfully inherited in subsequent generations in agriculturally important crop species is an interesting area for further investigation. Nature Comms. 10.1038/s41467-018-03289-7