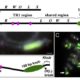
Loss of mCHH islands in maize chromomethylase and DDM1-type nucleosome remodeler mutants
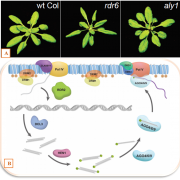
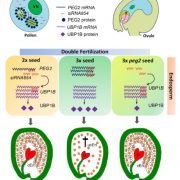
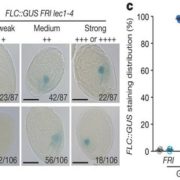
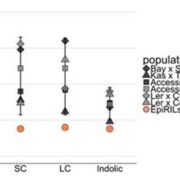
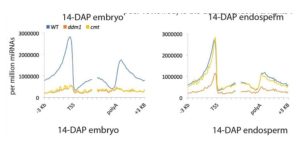 Plant DNA methylation in different sequence contexts is catalyzed by distinct types of methyltransferases. METHYLTRANSFERASE 1 (MET1) is responsible for CG methylation, CHROMOMETHYLASE (CMT1, 2, and 3) methylates CHG and CHH, while DOMAINS REARRANGED METHYLTRANSFERASE (DRM1 and 2) methylates in all three contexts through the RNA-directed DNA Methylation (RdDM) pathway. DECREASED DNA METHYLATION1 (DDM1) provides DNA methyltransferase access to heterochromatin to silence transposable elements together with the RdDM pathway. Single mutants of CMT (ZMET2 and ZMET5) and DDM1 (CHR101 and CHR106) in maize show some defects in DNA methylation, while double mutants of chr101 and chr106 and double mutants of zmet2 and zmet5 are nonviable. Arabidopsis mutants of the equivalent genes produce DNA methylation and small RNA changes, but little or no phenotypic consequences at the whole plant level. Fu et al. attempt to understand the relationship between RdDM and other forms of DNA methylation by looking at methylation and small RNA signatures produced by double knockouts of CMT (ZMET2 and ZMET5) and DDM1 (CHR101 and CHR106) in maize. The authors show that double mutants produce embryo and endosperm, and examine DNA methylation and small RNA changes in both tissues. In the developing embryo, there was a complete loss of 24nt siRNA and CHH methylation at CHH islands. 21nt and 22nt siRNA were gained in heterochromatin, although these 21-22nt siRNAs did not direct DNA methylation of the heterochromatic loci. In both double mutants, the dominant siRNA type shifted to 21 and 22nt, while 24nt siRNAs from terminal inverted repeat transposons were reduced eight-fold. The ddm1 and cmt mutants’ stronger effect on RdDM in maize may be attributed to its larger genome size compared to Arabidopsis. bioRxiv
Plant DNA methylation in different sequence contexts is catalyzed by distinct types of methyltransferases. METHYLTRANSFERASE 1 (MET1) is responsible for CG methylation, CHROMOMETHYLASE (CMT1, 2, and 3) methylates CHG and CHH, while DOMAINS REARRANGED METHYLTRANSFERASE (DRM1 and 2) methylates in all three contexts through the RNA-directed DNA Methylation (RdDM) pathway. DECREASED DNA METHYLATION1 (DDM1) provides DNA methyltransferase access to heterochromatin to silence transposable elements together with the RdDM pathway. Single mutants of CMT (ZMET2 and ZMET5) and DDM1 (CHR101 and CHR106) in maize show some defects in DNA methylation, while double mutants of chr101 and chr106 and double mutants of zmet2 and zmet5 are nonviable. Arabidopsis mutants of the equivalent genes produce DNA methylation and small RNA changes, but little or no phenotypic consequences at the whole plant level. Fu et al. attempt to understand the relationship between RdDM and other forms of DNA methylation by looking at methylation and small RNA signatures produced by double knockouts of CMT (ZMET2 and ZMET5) and DDM1 (CHR101 and CHR106) in maize. The authors show that double mutants produce embryo and endosperm, and examine DNA methylation and small RNA changes in both tissues. In the developing embryo, there was a complete loss of 24nt siRNA and CHH methylation at CHH islands. 21nt and 22nt siRNA were gained in heterochromatin, although these 21-22nt siRNAs did not direct DNA methylation of the heterochromatic loci. In both double mutants, the dominant siRNA type shifted to 21 and 22nt, while 24nt siRNAs from terminal inverted repeat transposons were reduced eight-fold. The ddm1 and cmt mutants’ stronger effect on RdDM in maize may be attributed to its larger genome size compared to Arabidopsis. bioRxiv