Boosting plant disease resistance through epigenetics
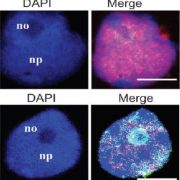
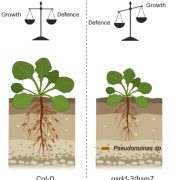
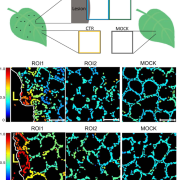
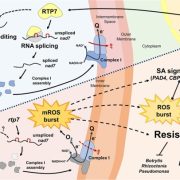
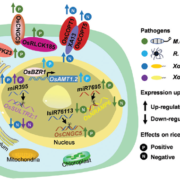
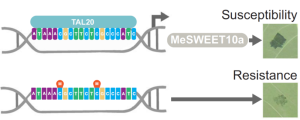 Increasing plant resistance to specific pathogens through genome editing is a very active branch of crop engineering. In a recent paper, Veley et al. propose to edit the epigenome to boost resistance to cassava bacterial blight by increasing the levels of DNA methylation at specific sites, while leaving the DNA sequence unchanged. The authors focus on the susceptibility gene MeSWEET10a, which is targeted by the bacterial effector TAL20. DNA binding by TAL20 is inhibited by methylation. The authors used an artificial zinc-finger (ZF) fused to Arabidopsis DEFECTIVE IN MERISTEM SILENCING 3 (DMS3) to specifically and stably methylate the effector binding element (EBE) targeted by TAL20 within the MaSWEET10a promoter. ZF-DMS3 transgenic plants have increased methylation levels at the MeSWEET10a EBE, but they show no phenotypic defects and their overall genome methylation levels are not affected. Moreover, MeSWEET10a expression, which increases upon infection in WT plants, was strongly reduced in ZF-DMS3 lines. Finally, the authors show that typical bacterial blight symptoms, such as water-soaking, were reduced in ZF-DMS3 plants, indicating that this strategy was successful in improving plant resistance to this disease. As more and more tools to edit the epigenome become available, modifying the DNA sequence may no longer be required to obtain the desired phenotype. (Summary by Laura Turchi @turchi_l) Nature Comms. 10.1038/s41467-022-35675-7.
Increasing plant resistance to specific pathogens through genome editing is a very active branch of crop engineering. In a recent paper, Veley et al. propose to edit the epigenome to boost resistance to cassava bacterial blight by increasing the levels of DNA methylation at specific sites, while leaving the DNA sequence unchanged. The authors focus on the susceptibility gene MeSWEET10a, which is targeted by the bacterial effector TAL20. DNA binding by TAL20 is inhibited by methylation. The authors used an artificial zinc-finger (ZF) fused to Arabidopsis DEFECTIVE IN MERISTEM SILENCING 3 (DMS3) to specifically and stably methylate the effector binding element (EBE) targeted by TAL20 within the MaSWEET10a promoter. ZF-DMS3 transgenic plants have increased methylation levels at the MeSWEET10a EBE, but they show no phenotypic defects and their overall genome methylation levels are not affected. Moreover, MeSWEET10a expression, which increases upon infection in WT plants, was strongly reduced in ZF-DMS3 lines. Finally, the authors show that typical bacterial blight symptoms, such as water-soaking, were reduced in ZF-DMS3 plants, indicating that this strategy was successful in improving plant resistance to this disease. As more and more tools to edit the epigenome become available, modifying the DNA sequence may no longer be required to obtain the desired phenotype. (Summary by Laura Turchi @turchi_l) Nature Comms. 10.1038/s41467-022-35675-7.