Terpenes in cannabis: Solving the puzzle of how to predict taste and smell
Marc-Sven Roell1
ORCID ID: 0000-0003-2714-8729
1Institute of Plant Biochemistry, Heinrich-Heine University Düsseldorf, Universitätsstrasse 1, 40225 Düsseldorf, Germany
Cannabis sativa (cannabis) is the cornerstone of the multi-billion-dollar legal marijuana industry. Cannabis plants are industrially used for fiber and oilseed production, but are primarily known for their resin, which is produced from glandular trichomes covering the surface of female flowers and is rich in cannabinoids and terpenes. The medicinal and psychoactive properties of cannabis depend on the total cannabinoid amount and the ratio of tetrahydrocannabinolic acid to cannabidiolic acid. However, fragrance and flavor are affected by terpene composition (Booth et al., 2017). To predict and design cannabis smell and taste to meet consumer demands, two milestones have to be reached. First, a comprehensive understanding of terpene composition is required, which can be achieved by using quantitative terpene profiling in existing cultivars. Second, the underlying molecular and biochemical mechanisms leading to these distinct profiles need to be understood.
In this issue of Plant Physiology, Booth et al. (2020) provide the framework for future breeding efforts to produce cannabis fragrance and flavor features demanded by consumers. Specifically, they analyzed terpene profiles of eight cannabis cultivars and characterized 13 novel cannabis terpene synthases. In plants, terpenes form a diverse group of hydrocarbon-based metabolites estimated to encompass thousands of different molecules (Pichersky and Raguso, 2018). Terpenes have diverse roles. They function as primary cellular components, e.g., as hormones or antioxidants, and they are indispensable for ecological interactions, e.g., signaling and defense against herbivores (Pichersky and Raguso, 2018). In cannabis, more than 100 different terpenes have been identified that define odor and flavor of different cultivars (Rothschild et al., 2005; Andre et al., 2016).
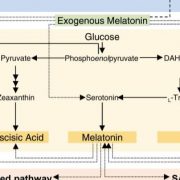
Terpene- and cannabinoid biosynthesis depend on the five-carbon building block isopentenyl pyrophosphate (IPP, Figure 1). IPP is produced by the plastidial methylerythritol phosphate (MEP) pathway and the cytosolic mevalonate (MEV) pathway. Metabolic fluxes within both pathways contribute to the substrate pools available for terpene synthases (TPSs). TPSs produce the diversity of cyclic and acyclic terpene core structures, using geranyl diphosphate or farnesyl diphosphate for monoterpene or sesquiterpene synthesis respectively (Figure 1).
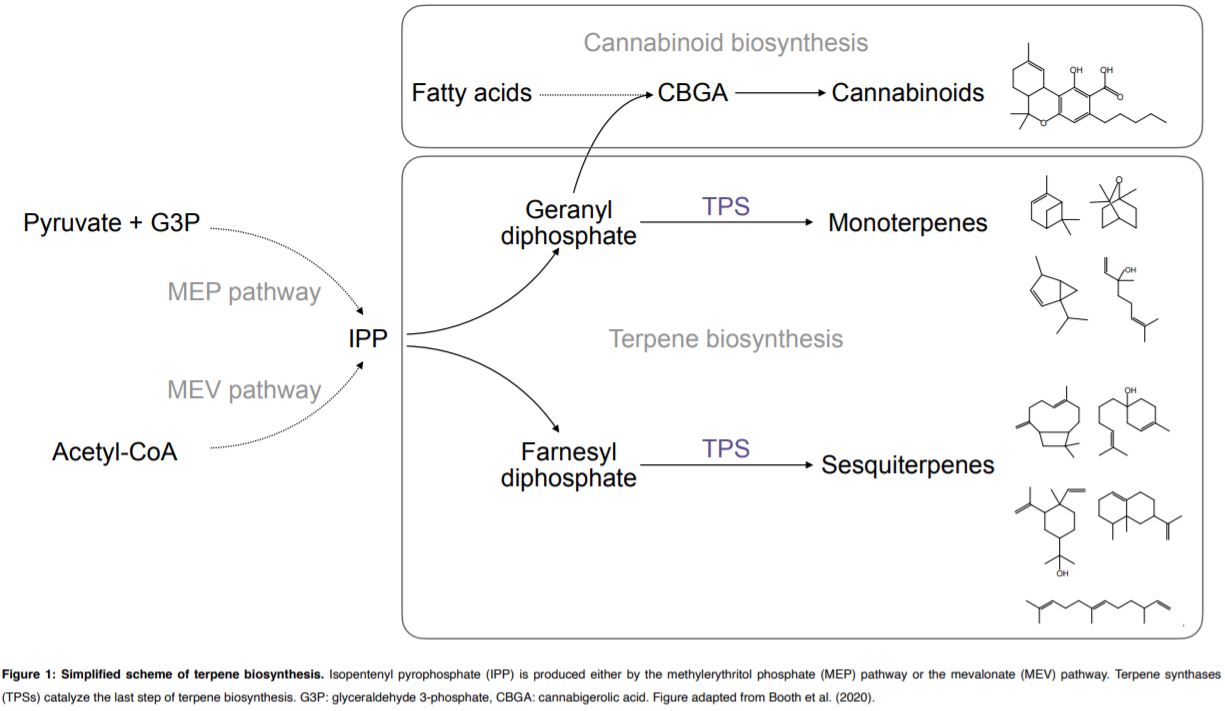 The accurate predicting and design of cannabis terpene profiles requires understanding of TPS, the key enzyme in terpene biosynthesis (Figure 1). Booth et al. identified 19 TPS gene models in the Purple Kush cannabis reference genome. TPS genes show multi-copy gene clustering, a common phenomenon previously observed for genes of the IPP and cannabinoid biosynthetic pathways (Taura et al., 2009). Foliar terpene profiling of eight cannabis cultivars revealed a total of 48 different terpenes with three monoterpenes (myrcene, α‐pinene, and limonene) and two sesquiterpenes (β‐caryophyllene and α‐humulene) present in all cultivars. In six selected cultivars, monoterpenes accumulated during the life cycle, in tissues including leaves, juvenile flowers and adult flowers. Using trichome transcriptome profiling, Booth et al. (2020) identified 33 TPS genes among the selected six cultivars. Further, 13 novel TPSs were biochemically characterized regarding product formation using both geranyl diphosphate and farnesyl diphosphate as substrates. Overall TPS specificity varied between the production of a single mono- or sesquiterpene to as many as 13 different sesquiterpenes produced by a single TPS enzyme.
The accurate predicting and design of cannabis terpene profiles requires understanding of TPS, the key enzyme in terpene biosynthesis (Figure 1). Booth et al. identified 19 TPS gene models in the Purple Kush cannabis reference genome. TPS genes show multi-copy gene clustering, a common phenomenon previously observed for genes of the IPP and cannabinoid biosynthetic pathways (Taura et al., 2009). Foliar terpene profiling of eight cannabis cultivars revealed a total of 48 different terpenes with three monoterpenes (myrcene, α‐pinene, and limonene) and two sesquiterpenes (β‐caryophyllene and α‐humulene) present in all cultivars. In six selected cultivars, monoterpenes accumulated during the life cycle, in tissues including leaves, juvenile flowers and adult flowers. Using trichome transcriptome profiling, Booth et al. (2020) identified 33 TPS genes among the selected six cultivars. Further, 13 novel TPSs were biochemically characterized regarding product formation using both geranyl diphosphate and farnesyl diphosphate as substrates. Overall TPS specificity varied between the production of a single mono- or sesquiterpene to as many as 13 different sesquiterpenes produced by a single TPS enzyme.
With these results, simple assumptions regarding TPS transcript abundance and terpene profiles can be made. However, spatiotemporal profiling of TPS transcript levels and terpene quantities will be necessary for more accurate predictions. Integrating the 13 TPSs characterized in this study with previously characterized TPS brings the total number to 30 known TPS across 14 different cannabis cultivars (Booth et al., 2017; Zager et al., 2019; Livingston et al., 2020). Harmonizing trichome transcriptomics tools, knowledge of TPS function and terpene profiling sets the framework for cannabis breeders to predictively shape and design terpene composition on demand.
Literature cited
Andre CM, Hausman J-F, Guerriero G (2016) Cannabis sativa: The plant of the thousand and one molecules. Front Plant Sci 7: 19
Booth JK, Page JE, Bohlmann J (2017) Terpene synthases from Cannabis sativa. PLoS ONE 12: e0173911
Livingston SJ, Quilichini TD, Booth JK, Wong DCJ, Rensing KH, Laflamme Yonkman J, Castellarin SD, Bohlmann J, Page JE, Samuels AL (2020) Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. Plant J 101: 37–56
Pichersky E, Raguso RA (2018) Why do plants produce so many terpenoid compounds? New Phytol 220: 692–702
Rothschild M, Bergström G, Wängberg SÅ (2005) Cannabis sativa: Volatile compounds from pollen and entire male and female plants of two variants, Northern Lights and Hawaian Indica. Botanical Journal of the Linnean Society 147: 387–397
Taura F, Tanaka S, Taguchi C, Fukamizu T, Tanaka H, Shoyama Y, Morimoto S (2009) Characterization of olivetol synthase, a polyketide synthase putatively involved in cannabinoid biosynthetic pathway. FEBS Lett 583: 2061–2066
Zager JJ, Lange I, Srividya N, Smith A, Markus Lange B (2019) Gene networks underlying cannabinoid and terpenoid accumulation in cannabis. Plant Physiol 180: 1877–1897