Systematic characterization of gene function in the photosynthetic alga Chlamydomonas reinhardtii (Nature Genetics)
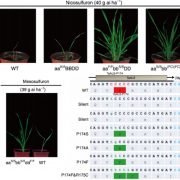
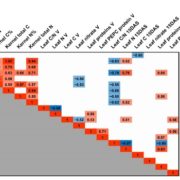
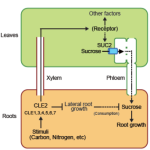
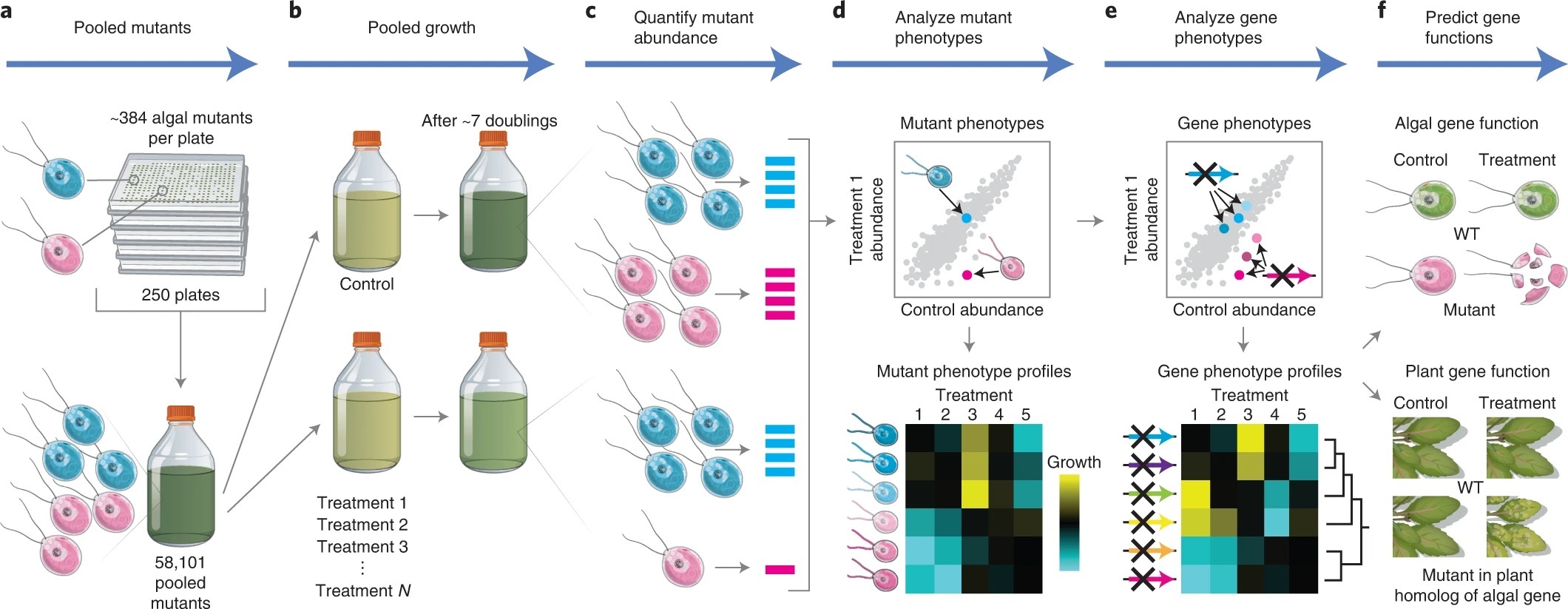 The green alga Chlamydomonas reinhardtii is a useful model system to study photosynthetic organisms, as this single-celled species allows for more high-throughput methods than in multicellular plants, and many conserved pathways of interest can be identified. Here, Fauser et al. demonstrate the value of these methods in a phenotypic screen of a previously generated barcoded insertion mutant library of Chlamydomonas. They used pooled cultures from a total of 58101 mutants covering 14695 genes, under 121 environmental and chemical stress conditions, using difference in growth as the assessed phenotype. They were able to make specific associations between genotypes and phenotypes and demonstrated the robustness of this method by confirming known genetic pathways, by clustering genes based on similar phenotypes. This also allowed them to characterise potential new players in various genetic pathways, such as the kinase MARS1 as playing a part in the activation of the chloroplast unfolded protein response upon DNA damage. They also identified three new genes in the defence pathway against actin inhibition by soil microorganisms (LAT5, LAT6, and LAT7) and showed conservation of function of these genes in Arabidopsis through mutant studies. Other characterised pathways include photosynthesis, CO2 concentrating mechanism, and cilia function, providing a useful resource for guiding gene characterisation in plants. (Summary by Jiawen Chen @jiaaawen ) Nature Genetics 10.1038/s41588-022-01052-9
The green alga Chlamydomonas reinhardtii is a useful model system to study photosynthetic organisms, as this single-celled species allows for more high-throughput methods than in multicellular plants, and many conserved pathways of interest can be identified. Here, Fauser et al. demonstrate the value of these methods in a phenotypic screen of a previously generated barcoded insertion mutant library of Chlamydomonas. They used pooled cultures from a total of 58101 mutants covering 14695 genes, under 121 environmental and chemical stress conditions, using difference in growth as the assessed phenotype. They were able to make specific associations between genotypes and phenotypes and demonstrated the robustness of this method by confirming known genetic pathways, by clustering genes based on similar phenotypes. This also allowed them to characterise potential new players in various genetic pathways, such as the kinase MARS1 as playing a part in the activation of the chloroplast unfolded protein response upon DNA damage. They also identified three new genes in the defence pathway against actin inhibition by soil microorganisms (LAT5, LAT6, and LAT7) and showed conservation of function of these genes in Arabidopsis through mutant studies. Other characterised pathways include photosynthesis, CO2 concentrating mechanism, and cilia function, providing a useful resource for guiding gene characterisation in plants. (Summary by Jiawen Chen @jiaaawen ) Nature Genetics 10.1038/s41588-022-01052-9