Sweet and Juicy: Identification and Origins of the Dry Alleles in Sorghum
Sorghum (Sorghum bicolor) is the fifth most important cereal crop globally and is considered to be the “camel among crops” due to its ability to flourish in low-nutrient soils and to withstand prolonged drought. Cultivated varieties are phenotypically and morphological diverse. Consequently, sorghum is grown as both a food source (grain sorghum) and as an emerging bioenergy crop (sweet sorghum) due to its juicy stems rich in soluble sugars (Mathur et al., 2017).
Sorghum stem juiciness is controlled by a single locus, Dry, and stalks are either dry and pithy (frequently found in grain sorghum) or juicy and sweet, where pithy is dominant to juicy. In a new study, Zhang et al. (2018) identify the Dry gene as encoding a plant-specific NAC transcription factor (TF) and present compelling evidence implicating Dry as a major regulator of secondary cell wall (SCW) biosynthesis.
Using a combination of classical map-based cloning and genome-wide association studies, the authors identified a narrow region on chromosome 6 containing four candidate genes. Detailed sequence comparisons and expression analysis of dry/pithy versus juicy lines revealed that the Dry locus encodes a NAC TF. Notably, juicy stem sorghum lines contain various loss-of-function mutations in the Dry exonic regions. Complementation analysis in an elite sweet sorghum line resulted in the pithy stem phenotype and a significantly reduced stem water content, confirming the identity of the Dry locus.
The authors also observed striking morphological and compositional differences in the cell walls of stems from sorghum lines containing the functional Dry allele (pithy) with those from lines carrying the null dry allele (juicy). The latter exhibited reduced cell wall thickness and widespread cell shape irregularities and collapse, likely resulting from altered cellulose/hemi-cellulose content. Indeed, orthologs of the Dry NAC TF in several other plant species have been shown to control SCW formation via a downstream regulatory network composed primarily of MYB TFs (Nakano et al., 2015). To confirm the involvement of DRY as a master regulator of SCW biosynthesis in sorghum, the authors examined the transcript levels of these TF orthologs together with known cellulose/hemicellulose biosynthetic genes in four sorghum lines via RNA-Seq analysis. Most genes showed high correlation (positive or negative) with the Dry/dry alleles, supporting the conservation of this SCW NAC/MYB regulatory network in sorghum. Functional conservation may also extend to other major cereals including maize (Zea mays) and rice (Oryza sativa), since they also contain Dry orthologs, pointing to a common ancestor.
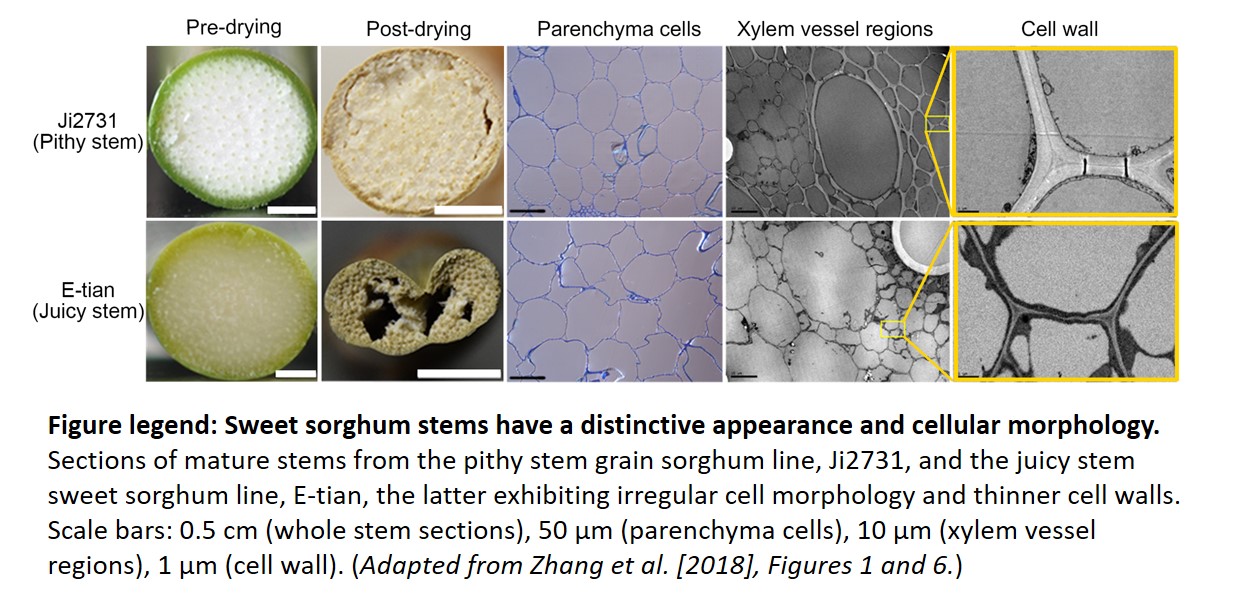
Sections of mature stems from the pithy stem grain sorghum line, Ji2731, and the juicy stem sweet sorghum line, E-tian, the latter exhibiting irregular cell morphology and thinner cell walls. Scale bars: 0.5 cm (whole stem sections), 50 µm (parenchyma cells), 10 µm (xylem vessel regions), 1 µm (cell wall). (Adapted from Zhang et al. [2018], Figures 1 and 6.)
The ancestry of sweet sorghum is not well defined, with evidence suggesting that it is of polyphyletic origin and therefore not easily separated from grain sorghum lines phylogenetically. Using a population genetics approach, the authors sequenced 241 sorghum accessions with a broad geographical distribution, including wild and cultivated lines, and identified several Dry gene haplotypes, indicative of a high degree of polymorphism. However, the complete lack of juicy haplotypes in wild sorghum strongly supports its occurrence as a result of relatively recent and intensive breeding selection, primarily for syrup production.
Although the locus for sorghum stem juiciness was discovered over 100 years ago, the gene responsible for this agriculturally and commercially important trait had not been identified previously. These findings are of major significance, since they pave the way for developing multipurpose sorghum (and potentially other cereal) varieties with juicy stems for biofuel production and other applications concomitant with high grain yield. In addition to characterizing the Dry gene, this study provides tantalizing historical insights into the domestication of sorghum.
REFERENCES
Mathur, S., Umakanth, A.V., Tonapi, V.A., Sharma, R., and Sharma, M.K. (2017). Sweet sorghum as biofuel feedstock: recent advances and available resources. Biotechnology for Biofuels: 1–19.
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N.A., and Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci 6: 9693–18.
Zhang, L.M. et al. (2018). Sweet Sorghum Originated through Selection of Dry, a Plant-specific NAC Transcription Factor Gene. Plant Cell doi: 10.1105/tpc.18.00313