Small Cells, Big Impact: RNA-Protein Complexes in a Cyanobacterium
Riediger et al. investigate the RNA-protein complexome of a photosynthetic cyanobacterium. The Plant Cell. https://bit.ly/3njpBn4
By Matthias Riediger and Wolfgang R. Hess
Background: Cyanobacteria are a unique group of bacteria with the ability to perform plant-like photosynthesis. Like their larger cousins, land plants and eukaryotic algae, they only require sunlight, inorganic nutrients and fairly stable environmental conditions to thrive. Unfortunately, these factors may change frequently and in unexpected ways. Accordingly, cyanobacteria have developed mechanisms to cope with such changes. One mechanism that is common to all forms of life is transcriptional gene regulation, but is often insufficient since it requires time. Therefore, posttranscriptional regulatory processes are often equally or even more important. Proteins that bind to and interact with regulatory small RNAs and other RNAs are crucially relevant but are seldom investigated in cyanobacteria.
Question: What are the cyanobacterial RNA-binding proteins involved in important post-transcriptional regulatory processes, which in contrast to other bacteria are not well understood? Posttranscriptional regulation may indeed differ fundamentally between cyanobacteria and non-photosynthetic bacteria.
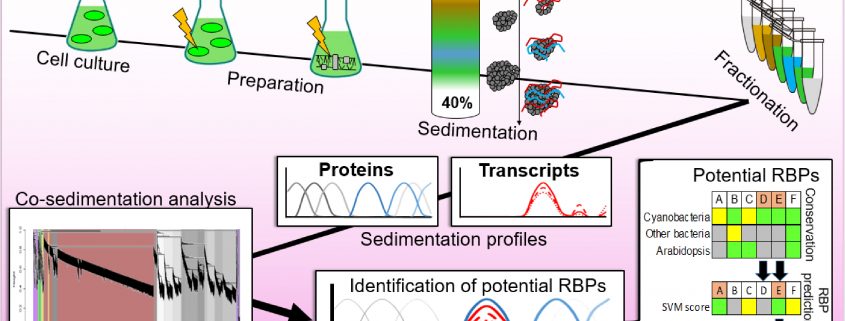
Findings: We present the global analysis of major stable RNA-protein complexes in a photosynthetic cyanobacterium. The intricate intracellular membrane systems and their propensity to associate with protein complexes differentiate cyanobacteria from other bacteria, but resemble the situation of plant photosynthetic cells, especially within chloroplasts. We investigated Synechocystis sp. PCC 6803, one of the most popular model cyanobacteria. We analyzed the sedimentation behavior of 65% of the 3,681 different proteins and 62.2% of the 4,251 different transcriptional units in a sucrose density gradient. We computed a support vector machine score for the prediction of RNA-binding proteins, focusing on proteins of unknown function that co-sediment with RNAs. Assuming that relevant proteins are more phylogenetically conserved, we looked for the presence of putative orthologs in a wider set of cyanobacteria, Arabidopsis thaliana, E. coli and Salmonella enterica and evaluated the synteny among 34 different cyanobacteria.
Next steps: We present a short list of RNA-binding protein candidates that can now be evaluated experimentally by characterizing respective mutants and their set of likely interacting RNAs. Our data not only provide crucial knowledge for this particular model organism but also constitute a global resource for cyanobacteria and photosynthetic organelles that can be accessed online at https://sunshine.biologie.uni-freiburg.de/GradSeqExplorer/.
Riediger M., Spät P., Bilger R., Macek B., Hess W.R. (2021). Analysis of a Photosynthetic Cyanobacterium Rich in Internal Membrane Systems via Gradient Profiling by Sequencing (Grad-seq). Plant Cell, https://bit.ly/3njpBn4