Saddle Up, Soybean Seed Pigments: Argonaute5 in Spatially Regulated Silencing of Chalcone Synthase Genes
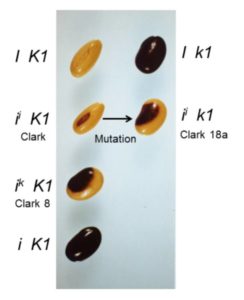 Most soybean seeds you see, whether in bins at the store, or in train cars as a commodity crop, have a yellow seed coat that may have only a tiny fleck of dark pigment at the hilum, where the seed attaches to the pod. The predominant yellow color results from silencing of chalcone synthase (CHS) genes by RNA interference (RNAi) due to the presence of I or ii alleles. The ii allele has an inverted duplication of CHS1, CHS3, and CHS4, causing the production of small interfering RNAs (siRNAs) that target CHS mRNAs and prevent pigment synthesis. Seeds carrying the rare ik allele have a larger region of pigment, producing a saddle pattern of dark purple pigment with black appearance on the yellow bean, more like a black-eyed pea. In a classic example of epistasis, the recessive k1 allele of the K1 locus overcomes the restriction of pigment to the hilum in ii plants: ii K1 soybeans have yellow seeds with a black hilum, but ii k1 soybeans have seeds with a black saddle (similar to ik K1 seeds), and I k1 soybeans have seeds that are entirely black.
Most soybean seeds you see, whether in bins at the store, or in train cars as a commodity crop, have a yellow seed coat that may have only a tiny fleck of dark pigment at the hilum, where the seed attaches to the pod. The predominant yellow color results from silencing of chalcone synthase (CHS) genes by RNA interference (RNAi) due to the presence of I or ii alleles. The ii allele has an inverted duplication of CHS1, CHS3, and CHS4, causing the production of small interfering RNAs (siRNAs) that target CHS mRNAs and prevent pigment synthesis. Seeds carrying the rare ik allele have a larger region of pigment, producing a saddle pattern of dark purple pigment with black appearance on the yellow bean, more like a black-eyed pea. In a classic example of epistasis, the recessive k1 allele of the K1 locus overcomes the restriction of pigment to the hilum in ii plants: ii K1 soybeans have yellow seeds with a black hilum, but ii k1 soybeans have seeds with a black saddle (similar to ik K1 seeds), and I k1 soybeans have seeds that are entirely black.
To identify the K1 locus, Cho et al. (2017) used RNA sequencing to compare ik K1 and ii k1 seeds, which have similar pigmentation patterns (figure). This showed that k1 has a 129-bp deletion in a gene encoding Argonaute5 (AGO5). AGO proteins bind small RNAs and participate in RNAi by different mechanisms, such as cleavage of mRNAs, inhibition of translation, sequestration of other small RNAs, and repression of transcription via DNA methylation (reviewed in Fang and Qi, 2016). Work in Arabidopsis thaliana and rice (Oryza sativa) implicated AGO5 homologs in the development of germ cells and defense against viruses (reviewed in Fang and Qi, 2016).
The AGO5 transcript from the k1 allele produces a spliced transcript that lacks all of exon 7; this transcript encodes a truncated protein missing one domain (and with partial deletion of another) that is essential for interaction with small RNAs. This change and the recessive nature of the k1 allele indicate that the AGO5 produced by this allele is non-functional. Re-sequencing of AGO5 in four independent lines with saddle patterns identified additional mutations in AGO5, supporting the hypothesis that loss of AGO5 function causes the epistatic effect with alleles of I.
To examine the mechanism by which the lack of AGO5 affects seed pigmentation, the authors dissected different regions (pigmented and un-pigmented) of the seed coat to compare siRNA abundance. The siRNAs targeting CHS are most abundant before the pigment develops; therefore, they dissected regions of the green seed coat based on position. High-throughput sequencing of small RNAs showed that the Williams cultivar, which has the ii K1 genotype, showed about 20-fold more siRNAs that target CHS loci in the region that will become yellow than in the hilum region that will become pigmented. Similarly, Clark 8a (ik K1 saddle pattern) and Clark 18a (ii k1 saddle pattern) seeds showed higher levels of siRNAs targeting CHS loci in the un-pigmented region. RNA-sequencing data (the same data used to identify K1 above) showed that the regions with high levels of CHS siRNAs had correspondingly low levels of CHS mRNAs. These non-pigmented regions also had the highest levels of AGO5 mRNA, and AGO5 mRNA levels were much lower in k1 seed coats, compared with K1. Intriguingly, of the twenty annotated AGO loci in the soybean genome, many were highly expressed in the seed coat and several, including AGO10, showed higher expression levels in the non-pigmented regions.
This interesting study showed that AGO5 affects the distribution of siRNAs targeting the CHS genes in the soybean seed coat. As the authors point out, many soybean AGOs likely function in the RNAi suppression of CHS genes in the seed coats. Moreover, elucidation of developmental factors that specify the spatial boundary and how the RNAi factors spatially regulate the levels of siRNAs, whether via effects on transcription, RNA cleavage, small RNA biogenesis, or another mechanism, remains an intriguing topic for future studies.
IN BRIEF by Jennifer Mach [email protected]
REFERENCES
Cho, Y.B., Jones, S.I., and Vodkin, L.O. (2017) Mutations in Argonaute5 Illuminate Epistatic Interactions of the K1 and I Loci Leading to Saddle Seed Color Patterns in Glycine max. Plant Cell 10.1105/tpc.17.00162.
Fang, X. and Qi, Y. (2016). RNAi in Plants: An Argonaute-Centered View. The Plant Cell 28: 272-285.



Leave a Reply
Want to join the discussion?Feel free to contribute!