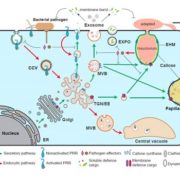
Roles for this ROP GTPase in subcellular and tissue-level patterning (Plant Cell)
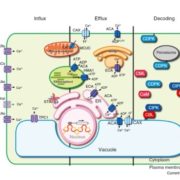
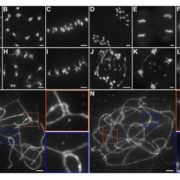
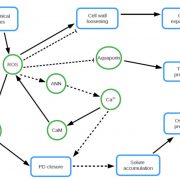
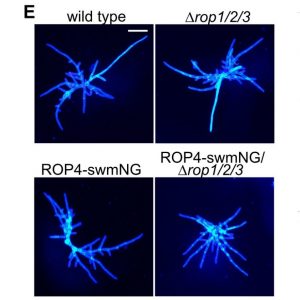 Rho-of-plants (ROP) proteins are master regulators of cell polarity within plants and influence cell wall deposition, tissue development, and several signaling processes. However, they are quite difficult to study: attempts to form a fluorescent ROP fusion protein have proven elusive, and there are multiple functionally redundant ROP family members in seed plants, whereas Physcomitrium patens contains only four ROP proteins. As ROPs are promiscuous and contain multiple interaction sites, choosing the “wrong” site to tag can (and does, as the authors of this paper show) negatively affect native protein function, generating inaccurate data. In this study, Cheng et al. used P. patens to create a mutant line containing null mutations of ROP1/2/3 and an edited ROP4 with a mNeonGreen “sandwich” tag in the middle of the sequence, generating a fluorescently-tagged ROP4 (referred to by the authors and further in this summary as “ROP4-swmNG”) that did not interfere with known interaction sites; was phenotypically similar to the WT; and sufficiently rescued ROP activity. Using this powerful tool, the authors were able to determine notable ROP4 functions. During protonemata development, ROP4-swmNG accumulated at the apical tip of actively growing cells, and their accumulation was shown to consistently predict the sites of cellular expansion. Further investigations suggested that the localization of ROP4-swmNG was maintained by both actin and microtubules, but specifically in the early stages of the cell cycle, hinting at a cell cycle function. Such studies are vital towards both inspirational tool development and further characterization of important plant growth components. (Summary by Benjamin Jin) Plant Cell 10.1105/tpc.20.00440
Rho-of-plants (ROP) proteins are master regulators of cell polarity within plants and influence cell wall deposition, tissue development, and several signaling processes. However, they are quite difficult to study: attempts to form a fluorescent ROP fusion protein have proven elusive, and there are multiple functionally redundant ROP family members in seed plants, whereas Physcomitrium patens contains only four ROP proteins. As ROPs are promiscuous and contain multiple interaction sites, choosing the “wrong” site to tag can (and does, as the authors of this paper show) negatively affect native protein function, generating inaccurate data. In this study, Cheng et al. used P. patens to create a mutant line containing null mutations of ROP1/2/3 and an edited ROP4 with a mNeonGreen “sandwich” tag in the middle of the sequence, generating a fluorescently-tagged ROP4 (referred to by the authors and further in this summary as “ROP4-swmNG”) that did not interfere with known interaction sites; was phenotypically similar to the WT; and sufficiently rescued ROP activity. Using this powerful tool, the authors were able to determine notable ROP4 functions. During protonemata development, ROP4-swmNG accumulated at the apical tip of actively growing cells, and their accumulation was shown to consistently predict the sites of cellular expansion. Further investigations suggested that the localization of ROP4-swmNG was maintained by both actin and microtubules, but specifically in the early stages of the cell cycle, hinting at a cell cycle function. Such studies are vital towards both inspirational tool development and further characterization of important plant growth components. (Summary by Benjamin Jin) Plant Cell 10.1105/tpc.20.00440