Review: Less is more, natural loss-of-function mutation is a strategy for adaptation (Plant Communications)
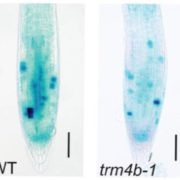
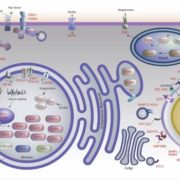
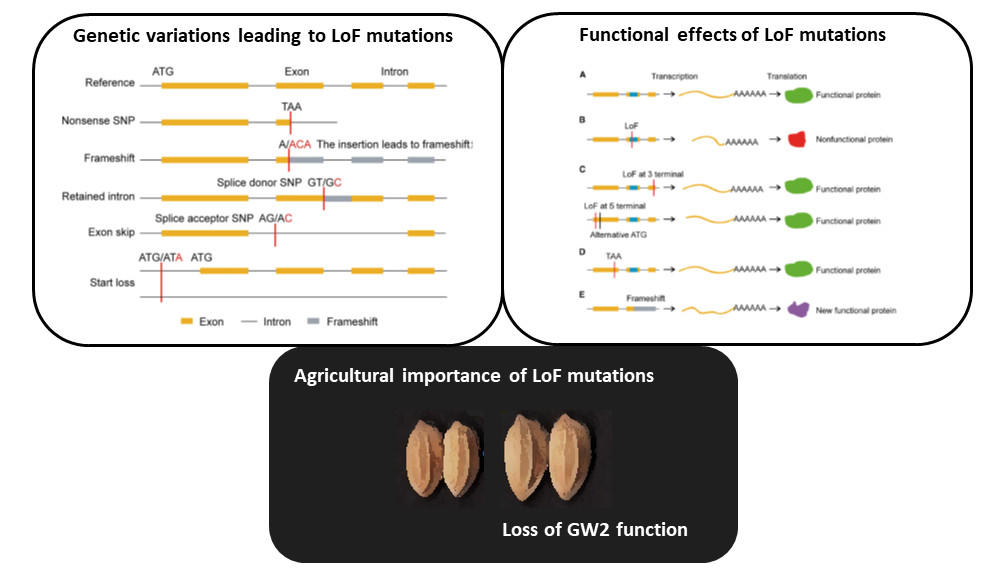 Gene gain through duplication and gene loss through loss-of-function (LoF) mutations determine genetic variation underlying diversification and adaptive evolution. In this review, Xu and Guo highlight the importance of LoF mutations in the evolutionary success of several organisms as revealed by advancements in whole-genome sequencing. The authors focused on the genome-wide distribution of LoF variants in natural populations. Many deleterious LoF mutations show low allele frequencies and tend to accumulate in redundant genes belonging to large families (as paralogs can take over their function). Similar genome-wide analyses also provide information on the “essential gene set” of an organism: absence of deleterious LoF variants at a given gene suggests its crucial role for survival. Nevertheless, not all the LoF mutations lead to complete knockouts because the functional effect also depends on their location. Moreover, LoF mutations could generate truncated proteins with a dominant-negative effect or new functional proteins with similar features. In essence, LoF variants seem to have a great impact on adaptation of the species to new environments. Supporting evidence comes from genetic studies in Petunia: LoF mutations in genes controlling flower scent and color determined changes in pollinator attraction. The authors suggest boosting this approach in crops: novel LoF mutations could be employed as valuable source of genetic variation in breeding programs. For example, LoF variants in Grain Width 2 (GW2) have already been used to increase grain yield in rice. To conclude, gene loss could also be beneficial as proposed by the LESS IS MORE hypothesis. (Summary and image adaptation by Michela Osnato @michela_osnato) Plant Communications 10.1016/j.xplc.2020.100103
Gene gain through duplication and gene loss through loss-of-function (LoF) mutations determine genetic variation underlying diversification and adaptive evolution. In this review, Xu and Guo highlight the importance of LoF mutations in the evolutionary success of several organisms as revealed by advancements in whole-genome sequencing. The authors focused on the genome-wide distribution of LoF variants in natural populations. Many deleterious LoF mutations show low allele frequencies and tend to accumulate in redundant genes belonging to large families (as paralogs can take over their function). Similar genome-wide analyses also provide information on the “essential gene set” of an organism: absence of deleterious LoF variants at a given gene suggests its crucial role for survival. Nevertheless, not all the LoF mutations lead to complete knockouts because the functional effect also depends on their location. Moreover, LoF mutations could generate truncated proteins with a dominant-negative effect or new functional proteins with similar features. In essence, LoF variants seem to have a great impact on adaptation of the species to new environments. Supporting evidence comes from genetic studies in Petunia: LoF mutations in genes controlling flower scent and color determined changes in pollinator attraction. The authors suggest boosting this approach in crops: novel LoF mutations could be employed as valuable source of genetic variation in breeding programs. For example, LoF variants in Grain Width 2 (GW2) have already been used to increase grain yield in rice. To conclude, gene loss could also be beneficial as proposed by the LESS IS MORE hypothesis. (Summary and image adaptation by Michela Osnato @michela_osnato) Plant Communications 10.1016/j.xplc.2020.100103