More than Window Dressing: Revealing 5-Methylcytocine Patterns that Decorate Arabidopsis RNA
IN BRIEF by Jennifer Lockhart [email protected]
DNA is sculpted by several types of epigenetic modifications with profound effects on gene expression, development, and stress responses. Much less is known about the more than 100 chemical modifications shaping plant RNA, a topic explored in the newly emerging field of epitranscriptomics (reviewed in Burgess et al., 2016). These post-translational modifications can occur on the RNA base itself or on the ribose sugar backbone of the delicate RNA molecule. One such modification, the conversion of cytosine residues to 5-methylcytocine (m5C), affects the stability and translation of tRNA and the processing, structure, and translation of rRNA in many organisms. Due to technical challenges, much less is known about the roles of m5C in low-abundance RNAs, such as mRNAs and various non-coding RNAs (ncRNAs). The enzymes that mediate m5C methylation have also remained elusive.
David et al. (2017) attacked this issue through transcriptome-wide, single nucleotide resolution analysis of m5C patterns in Arabidopsis thaliana. Their study combined RNA bisulfite conversion (Schaefer et al., 2009) with second-generation Illumina sequencing (bsRNA-seq) of silique, shoot, and root tissue. Treating RNA with bisulfite selectively deaminates cytosine (but not m5C) to uracil, allowing these converted cytosine residues (and hence, the remaining m5C sites) to be detected by Illumina sequencing. By aligning the Illumina reads to the Arabidopsis transcriptome, the authors identified hundreds of novel m5C sites on many types of RNA in all three tissue types, as well as tissuespecific patterns of m5C modification. Candidate m5C sites were tested using RNA bisulfite amplicon sequencing (nextgeneration sequencing of targeted PCR amplicons derived from bisulfite-treated total RNA) and anti-m5C immunoprecipitation, confirming the robustness of bsRNA-seq.
The focus turned to the Arabidopsis tRNA-specific methyltransferase 4b (trm4b) mutant, as RNA methyltransferases can catalyze m5C modification of mRNA and other types of ncRNAs. Transcriptome-wide bsRNA-seq of RNA from wild-type and trm4b silique, seedling shoot, and root tissue revealed numerous sites with no or reduced methylation compared to wild type, suggesting that TRM4B mediates the formation of many m5C sites on diverse classes of RNA across all three tissue types. Indeed, overexpressing TRM4B led to a 3-and 124-fold increase in methylation percentage, specifically at TRM4B-dependent m5C sites. Furthermore, a 50-nucleotide sequence surrounding the m5C sites is sufficient to recruit TRM4B for m5C modification, as determined by a transient expression assay in Nicotiana benthamiana leaves.
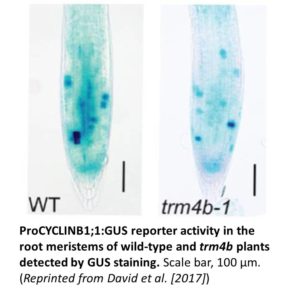 While shoot and inflorescence growth appear normal in trm4b, its primary roots are significantly shorter than wild type, likely due to reduced cell division in the meristem. Indeed, a marked reduction in mitotic activity was evident in trm4b roots compared to wild type, which was detected using a ProCYCLIN B1;1-GUS reporter line (to reveal mitotic activity) crossed into the wild-type versus trm4b background (see figure). Since m5C methyltransferases are important oxidative stress response regulators in several species, the authors investigated the response of trm4b to this stress. The mutant showed increased sensitivity to oxidative stress but not to salt stress, suggesting that TRM4B mediates the oxidative stress response in Arabidopsis. The mutant phenotypes could be due to altered m5C levels in tRNAs (leading to reduced tRNA stability) rather than in the identified mRNAs, a promising area for future study. Oxidative stress-related genes are activated in the mutant, even under control conditions, suggesting the short root phenotype of trm4b might be mediated by the partial activation of oxidative stress pathways.
While shoot and inflorescence growth appear normal in trm4b, its primary roots are significantly shorter than wild type, likely due to reduced cell division in the meristem. Indeed, a marked reduction in mitotic activity was evident in trm4b roots compared to wild type, which was detected using a ProCYCLIN B1;1-GUS reporter line (to reveal mitotic activity) crossed into the wild-type versus trm4b background (see figure). Since m5C methyltransferases are important oxidative stress response regulators in several species, the authors investigated the response of trm4b to this stress. The mutant showed increased sensitivity to oxidative stress but not to salt stress, suggesting that TRM4B mediates the oxidative stress response in Arabidopsis. The mutant phenotypes could be due to altered m5C levels in tRNAs (leading to reduced tRNA stability) rather than in the identified mRNAs, a promising area for future study. Oxidative stress-related genes are activated in the mutant, even under control conditions, suggesting the short root phenotype of trm4b might be mediated by the partial activation of oxidative stress pathways.
It is currently unclear if m5C modifications of mRNAs or ncRNAs other than tRNAs are biologically significant rather than random decorations, but this is likely the case. The mapping of m5C in Arabidopsis provides the necessary tools to address this intriguing issue.
REFERENCES
Burgess, A., David, R., and Searle, I.R. (2016) Deciphering the epitranscriptome: A green perspective. Journal of Integrative Plant Biology 58: 822-835.
David, R., Burgess, A., Parker, B., Li, J., Pulsford, K., Sibbritt, T., Preiss, T., and Robert Searle, I.R. (2017) Transcriptomewide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and non-coding RNAs. Plant Cell 29: 10.1105/tpc.16.00751
Schaefer M, Pollex T, Hanna K, Lyko F. (2009). RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Research 37: e12.




Leave a Reply
Want to join the discussion?Feel free to contribute!