Review: Deep learning for plant genomics and crop improvement (Curr. Opin. Plant Biol.)
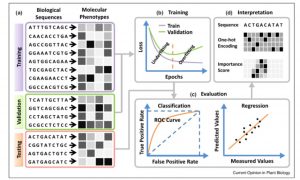 One of the goals of plant science is to use the molecular phenotype (genome, transcriptome, proteome) to predict the whole-plant phenotype. Deep learning approaches can potentially begin to do this, starting with a training dataset, and testing it with a validation dataset. Wang et al. review advances in towards this goal, particularly those that employ deep learning and neural networks, such as Convolutional Neural Networks (CNNs). The authors define different types of neural networks, and how they can be optimized for example to identify single nucleotide effects on traits, or on transcription factor binding. They also describe methods that have been developed to predict protein structure or protein-protein interactions. The authors discuss the possibilities and pitfalls of adopting deep learning models developed for animal or human genomes to plants, as well as plant-specific challenges such as polyplody. Plants’ phenotypic sensitivity to the environment adds additional complexity. The authors conclude that deep learning is a “promising approach for genome-wide identification of deleterious and adaptive variants, a prerequisite for editing-based genetic improvement of crops in future agriculture.” (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2019.12.010
One of the goals of plant science is to use the molecular phenotype (genome, transcriptome, proteome) to predict the whole-plant phenotype. Deep learning approaches can potentially begin to do this, starting with a training dataset, and testing it with a validation dataset. Wang et al. review advances in towards this goal, particularly those that employ deep learning and neural networks, such as Convolutional Neural Networks (CNNs). The authors define different types of neural networks, and how they can be optimized for example to identify single nucleotide effects on traits, or on transcription factor binding. They also describe methods that have been developed to predict protein structure or protein-protein interactions. The authors discuss the possibilities and pitfalls of adopting deep learning models developed for animal or human genomes to plants, as well as plant-specific challenges such as polyplody. Plants’ phenotypic sensitivity to the environment adds additional complexity. The authors conclude that deep learning is a “promising approach for genome-wide identification of deleterious and adaptive variants, a prerequisite for editing-based genetic improvement of crops in future agriculture.” (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2019.12.010