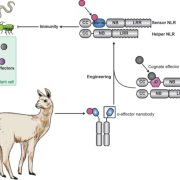
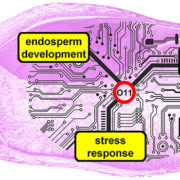
Review. Decoding resilience: Ecology, regulation, and evolution of biosynthetic gene clusters
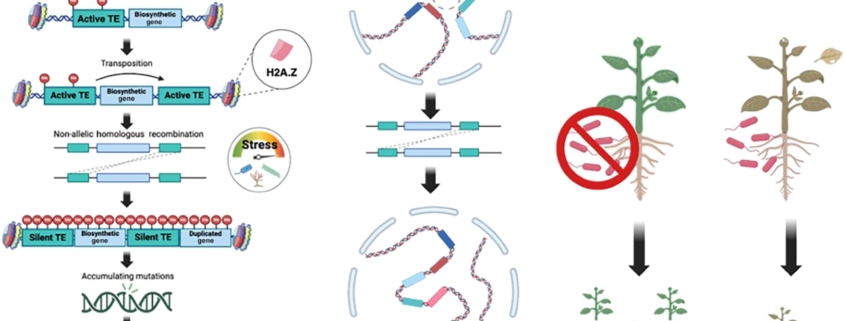
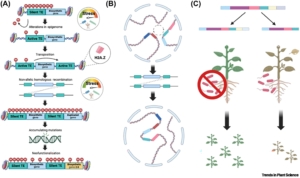 Although clusters of functionally related genes are common in prokaryotes, until recently it was thought that they were not a feature of eukaryotic genomes. However, several studies have identified biosynthetic gene clusters (BGCs) in plants. Many of these gene clusters include sets of enzymes that act sequentially in the production of specialized metabolites, such as defense or signaling molecules, enabling rapid and cost-effective production of the compound. A new review by Cawood and Ton discusses the function and regulation of these BGCs and speculates on how they are formed. Unlike prokaryotes, which have polycistronic BGCs (with many proteins encoded by a single mRNA), BGCs in eukaryotes are monocistronic, yet closely co-regulated, raising the question of how. Many studies suggest a role for histone modifications and histone variants (e.g., H2A.Z) in BGC co-regulation. There is also evidence for 3D chromatin topology and the formation of topologically associated domains (TADs) in BGC regulation. For example, in one case a chromatin loop structure leads to five BGC promoters simultaneously interacting with transcription factors and coactivators. Finally, the review turns to the question of how these BGCs were formed. It is generally assumed that they are derived from gene duplication and neofunctionalization, but the details of how this took place are not known. The authors describe a model in which stress induces activation of transposable elements, leading to gene duplication, exon shuffling, and gene fusion, providing opportunities for neofunctionalization. The authors also postulate that topologically associated domains could lead to related genes repositioning into BGCs. Finally, they note that in addition to positive selection of functional BGCs, there can be negative selection against incomplete BGCs due to the accumulation of potentially harmful metabolites. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci 10.1016/j.tplants.2024.09.008
Although clusters of functionally related genes are common in prokaryotes, until recently it was thought that they were not a feature of eukaryotic genomes. However, several studies have identified biosynthetic gene clusters (BGCs) in plants. Many of these gene clusters include sets of enzymes that act sequentially in the production of specialized metabolites, such as defense or signaling molecules, enabling rapid and cost-effective production of the compound. A new review by Cawood and Ton discusses the function and regulation of these BGCs and speculates on how they are formed. Unlike prokaryotes, which have polycistronic BGCs (with many proteins encoded by a single mRNA), BGCs in eukaryotes are monocistronic, yet closely co-regulated, raising the question of how. Many studies suggest a role for histone modifications and histone variants (e.g., H2A.Z) in BGC co-regulation. There is also evidence for 3D chromatin topology and the formation of topologically associated domains (TADs) in BGC regulation. For example, in one case a chromatin loop structure leads to five BGC promoters simultaneously interacting with transcription factors and coactivators. Finally, the review turns to the question of how these BGCs were formed. It is generally assumed that they are derived from gene duplication and neofunctionalization, but the details of how this took place are not known. The authors describe a model in which stress induces activation of transposable elements, leading to gene duplication, exon shuffling, and gene fusion, providing opportunities for neofunctionalization. The authors also postulate that topologically associated domains could lead to related genes repositioning into BGCs. Finally, they note that in addition to positive selection of functional BGCs, there can be negative selection against incomplete BGCs due to the accumulation of potentially harmful metabolites. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci 10.1016/j.tplants.2024.09.008