R-Loop Mediated trans Action of the APOLO Long Noncoding RNA
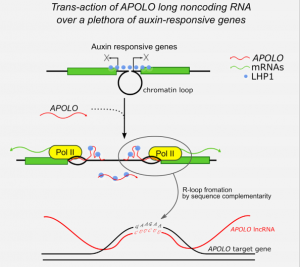
It was shown that the Arabidopsis intergenic long noncoding RNA (LncRNA) APOLO (AUXIN-REGULATED PROMOTER LOOP) modulates in cis its neighbor gene PID forming a chromatin loop. However, it remained unknown whether it can act also in trans modulating the expression of other genes. Now, Ariel et al. performed chromatin isolation by APOLO-RNA purification followed by DNA-seq (ChIRP-seq) and RNA-seq of overexpressing APOLO lines in order to identify potential targets in the Arabidopsis genome. They found a group of “bona fide” targets also related to auxin response. These target loci are also enriched in the binding of the Polycomb Repressive Complex 1 (PRC1) component LHP1 and the formation of DNA-RNA duplex (R-loops). They propose a model where APOLO is mediating the formation of R-loops and decoying the PRC1 complex over target loci to modulate gene expression. In order to demonstrate that, they performed DRIP-qPCR and developed a novel method named RNA isolation by DNA purification (RIDP) to purify a specific DNA locus to further isolate RNA and quantify R-loop dependent APOLO presence using RNAseH. This paper shows that lncRNA can shape the three-dimensional organization of genetic information and are a promising source of distal transcriptional regulators in plants. (Summary by Facundo Romani) Molecular Cell 10.1016/j.molcel.2019.12.015