Plant Science Research Weekly: September 6th
Review: Metabolite control of translation by conserved peptide uORFs: The ribosome as a metabolite multi-sensor
 Not all mRNAs are translated equally. Between 25–50% of eukaryotic mRNAs have an upstream open reading frame (uORF) that affects translation of the main ORF (mORF). Usually the presence of an uORF inhibits translation, but under some conditions the ribosome can overlook the uORF or reinitiate translation and precede to translate the mORF. Many factors including the uORF sequence determine how it will affect translation. CPuORFs encode conserved peptides with distinct affects on translation, often in a metabolite-dependent fasion. Check out the fine review by van der Horst et al. for more on how, through the CPuORFs, the ribosome acts as a metabolite multi-sensor. (Summary by Mary Williams) Plant Physiol 10.1104/pp.19.00940
Not all mRNAs are translated equally. Between 25–50% of eukaryotic mRNAs have an upstream open reading frame (uORF) that affects translation of the main ORF (mORF). Usually the presence of an uORF inhibits translation, but under some conditions the ribosome can overlook the uORF or reinitiate translation and precede to translate the mORF. Many factors including the uORF sequence determine how it will affect translation. CPuORFs encode conserved peptides with distinct affects on translation, often in a metabolite-dependent fasion. Check out the fine review by van der Horst et al. for more on how, through the CPuORFs, the ribosome acts as a metabolite multi-sensor. (Summary by Mary Williams) Plant Physiol 10.1104/pp.19.00940
Fellowship of the rings: a saga of strigolactones and other small signals
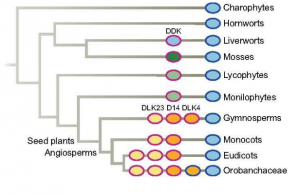 Strigolactones are small signal molecules synthesized by plants. In the past few years, many studies highlighting the importance of this emerging phytohormone have been published. Strigolactones play important roles as a hormonal signals in plants and for mycorrhizal fungi interactions, they are present in all land plants, and the signaling pathways seem to be conserved. But what do we really know about strigolactones? Many related small molecules (like karrikins) present similar biological activity and overlapping features, showing that there is not a simple pathway but an array of related signaling pathways. Here, Machin, Hamon-Josse & Bennett, make a big effort to review the recent literature on this topic and put it into context of strigolactone complexity and evolution. Among different topics, the authors discuss chemical properties, biosynthesis, and ligand-receptor specificity of strigolactones-related compounds. Downstream proteins involved in their signaling pathway from perception to transcriptional changes are also well covered. The promiscuity of the pathway is presented as an important feature that might shape their evolution to control different developmental processes in both plant and fungi. Finally, the authors also highlight open questions that still require further research. (Summary by Facundo Romani) New Phytol. 10.1111/nph.16135)
Strigolactones are small signal molecules synthesized by plants. In the past few years, many studies highlighting the importance of this emerging phytohormone have been published. Strigolactones play important roles as a hormonal signals in plants and for mycorrhizal fungi interactions, they are present in all land plants, and the signaling pathways seem to be conserved. But what do we really know about strigolactones? Many related small molecules (like karrikins) present similar biological activity and overlapping features, showing that there is not a simple pathway but an array of related signaling pathways. Here, Machin, Hamon-Josse & Bennett, make a big effort to review the recent literature on this topic and put it into context of strigolactone complexity and evolution. Among different topics, the authors discuss chemical properties, biosynthesis, and ligand-receptor specificity of strigolactones-related compounds. Downstream proteins involved in their signaling pathway from perception to transcriptional changes are also well covered. The promiscuity of the pathway is presented as an important feature that might shape their evolution to control different developmental processes in both plant and fungi. Finally, the authors also highlight open questions that still require further research. (Summary by Facundo Romani) New Phytol. 10.1111/nph.16135)
A Rubisco-binding protein is required for normal pyrenoid number and starch sheath morphology in Chlamydomonas reinhardtii
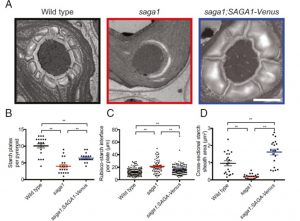 In most eukaryotic algae, carbon fixation takes place in an organelle within an organelle, the pyrenoid inside of the chloroplast. Besides being functionally very important, pyrenoids are interesting because they are what is called a phase-separated structure, that is they are not membrane enclosed; rather, they are usually surrounded by starch granules known as starch plates. Itakura et al. identified a Chlamydomonas mutant that forms multiple, abnormal pyrenoids, enclosed in abnormal starch plates. They found that the protein affected in this mutant, SAGA1, is a starch- and Rubisco-binding protein. Based on this and other work, the authors propose a model in which SAGA1 affects starch plate formation, and when the surface area determined by starch plates is excessive, the pyrenoid forms as multiple structures. They also suggest that surface area may be important in morphology of other phase-separated organelles. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1904587116
In most eukaryotic algae, carbon fixation takes place in an organelle within an organelle, the pyrenoid inside of the chloroplast. Besides being functionally very important, pyrenoids are interesting because they are what is called a phase-separated structure, that is they are not membrane enclosed; rather, they are usually surrounded by starch granules known as starch plates. Itakura et al. identified a Chlamydomonas mutant that forms multiple, abnormal pyrenoids, enclosed in abnormal starch plates. They found that the protein affected in this mutant, SAGA1, is a starch- and Rubisco-binding protein. Based on this and other work, the authors propose a model in which SAGA1 affects starch plate formation, and when the surface area determined by starch plates is excessive, the pyrenoid forms as multiple structures. They also suggest that surface area may be important in morphology of other phase-separated organelles. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1904587116
To be in Petri or in soil, that is the question
 Frequently, research on root growth in Arabidopsis has been carried out in transparent Petri dishes, although in the natural environment the soil-buried root actually grows in complete darkness. It is an essential question to check if experimental results reflect the natural growth of plants. It is well-known that when phosphate is deficient in a Petri dish, the primary root growth of Arabidopsis is inhibited. Recently Zheng et al. reported the molecular mechanism of how phosphate deficiency reshapes primary root growth in Petri dishes. The breakthrough is that actually the inhibited growth of primary root is just a phenotype of Arabidopsis to adapt to the blue light illumination of Petri. When grown in soil the primary root can elongate without inhibition. This discovery is not only of biological importance, but also raise a question whether the previous results obtained from Petri dish-grown Arabidopsis seedlings should be reevaluated. (Summary by Nanxun Qin) Molecular Plant 10.1016/j.molp.2019.08.001
Frequently, research on root growth in Arabidopsis has been carried out in transparent Petri dishes, although in the natural environment the soil-buried root actually grows in complete darkness. It is an essential question to check if experimental results reflect the natural growth of plants. It is well-known that when phosphate is deficient in a Petri dish, the primary root growth of Arabidopsis is inhibited. Recently Zheng et al. reported the molecular mechanism of how phosphate deficiency reshapes primary root growth in Petri dishes. The breakthrough is that actually the inhibited growth of primary root is just a phenotype of Arabidopsis to adapt to the blue light illumination of Petri. When grown in soil the primary root can elongate without inhibition. This discovery is not only of biological importance, but also raise a question whether the previous results obtained from Petri dish-grown Arabidopsis seedlings should be reevaluated. (Summary by Nanxun Qin) Molecular Plant 10.1016/j.molp.2019.08.001
Phosphorylation-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth
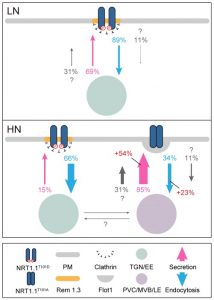 Nitrate is an important plant nutrient and multiple transporters have been identified in different plant species. NRT1.1 (NITRATE TRANSPORTER 1.1) belonging to the MFS (Major Facilitator Superfamily) has been shown to transport nitrate and auxin (IAA, Indole 3-acetic acid). This dual affinity transporter can switch from low- to high-nitrate affinity through differential phosphorylation of a threonine residue, Thr101. In this paper, Zhang et al. identified how this phosphorylation status can alter the transporter dynamics and thereby regulate lateral root growth. The authors altered the amino acid at position 101, to alter the phosphorylation status and found that the localization of the protein at the plasma membrane is altered. Non-phosphorylatable 101 residue led to oligomerization of the NRT1.1 transporter in the plasma membrane and altered auxin flux. This resulted in reduced lateral root growth in comparison to the plants expressing a phosphomimetic residue that can undergo regular phosphorylation. Through this work, NRT1.1 transporter regulation in response to phosphorylation of Thr101 has been shown to influence lateral root growth. (Summary by Suresh Damodaran) Plant Physiol. 10.1104/pp.19.00346
Nitrate is an important plant nutrient and multiple transporters have been identified in different plant species. NRT1.1 (NITRATE TRANSPORTER 1.1) belonging to the MFS (Major Facilitator Superfamily) has been shown to transport nitrate and auxin (IAA, Indole 3-acetic acid). This dual affinity transporter can switch from low- to high-nitrate affinity through differential phosphorylation of a threonine residue, Thr101. In this paper, Zhang et al. identified how this phosphorylation status can alter the transporter dynamics and thereby regulate lateral root growth. The authors altered the amino acid at position 101, to alter the phosphorylation status and found that the localization of the protein at the plasma membrane is altered. Non-phosphorylatable 101 residue led to oligomerization of the NRT1.1 transporter in the plasma membrane and altered auxin flux. This resulted in reduced lateral root growth in comparison to the plants expressing a phosphomimetic residue that can undergo regular phosphorylation. Through this work, NRT1.1 transporter regulation in response to phosphorylation of Thr101 has been shown to influence lateral root growth. (Summary by Suresh Damodaran) Plant Physiol. 10.1104/pp.19.00346
A CLE–SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza
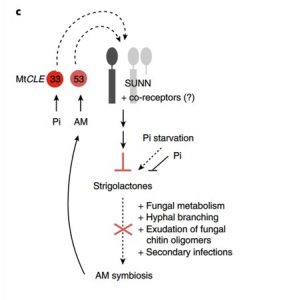 The symbiosis between plants and arbuscular mycorrhizal fungi is conditional. Plants that are limited for phosphate release strigolactones into the soil, promoting changes in the fungi that facilitate the symbiosis. Müller et al. found that a Medicago gene encoding a CLE peptide, MtCLE53, is induced upon root colonization. When this gene is ectopically overexpressed, plants resist colonization, and genes involved in strigolactone biosynthesis are downregulated. A gene encoding a receptor-like kinase, SUNN, was also upreagulated in MtCLE53-overexpressing plants. In sunn mutant plants, strigolactone biosynthesis is not downregulated in respones to MtCLE53-overexpression. Thus, a negative-feedback loop for symbiosis involving CLE53-SUNN and strigolactones has been identified. (Summary by Mary Williams) Nature Plants 10.1038/s41477-019-0501-1
The symbiosis between plants and arbuscular mycorrhizal fungi is conditional. Plants that are limited for phosphate release strigolactones into the soil, promoting changes in the fungi that facilitate the symbiosis. Müller et al. found that a Medicago gene encoding a CLE peptide, MtCLE53, is induced upon root colonization. When this gene is ectopically overexpressed, plants resist colonization, and genes involved in strigolactone biosynthesis are downregulated. A gene encoding a receptor-like kinase, SUNN, was also upreagulated in MtCLE53-overexpressing plants. In sunn mutant plants, strigolactone biosynthesis is not downregulated in respones to MtCLE53-overexpression. Thus, a negative-feedback loop for symbiosis involving CLE53-SUNN and strigolactones has been identified. (Summary by Mary Williams) Nature Plants 10.1038/s41477-019-0501-1
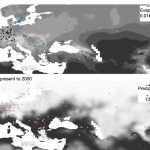
Natural selection on the Arabidopsis thaliana genome in present and future climates
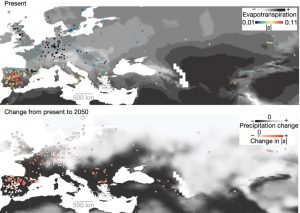 The rapidly changing climate will have profound effects on Earth’s ecosystems, but as yet it is difficult to determine exactly what these effects will be. Exposito-Alonso et al. have set up a large experiment to try to identify how a population’s genetic diversity will enable it to survive a future climate, by growing more than 500 natural accessions of Arabidopsis in Spain and Germany under different density and rainfall conditions. In the milder German conditions, representatives of all accessions survived and reproduced, but in the harsher Spanish conditions only 193 of the 517 accessions survived. They then examined how natural selection affected allele frequencies. Interestingly, many of the alleles that were under positive selection in the harsh conditions are under negative selection in the mild conditions (a phenomenon known as antagonistic pleiotropy). Starting with their data and the data from similar experiments, the authors built genome-wide environment selection (GWES) models to predict natural selection at various sites, essentially simulating field experiments. The models indicate that at the edges of Arabidopsis’ range, populations are at evolutionary risk. The authors point out that this proof-of-concept experiment can be adapted to design conservation strategies for at-risk species. (Summary by Mary Williams) Nature 10.1038/s41586-019-1520-9
The rapidly changing climate will have profound effects on Earth’s ecosystems, but as yet it is difficult to determine exactly what these effects will be. Exposito-Alonso et al. have set up a large experiment to try to identify how a population’s genetic diversity will enable it to survive a future climate, by growing more than 500 natural accessions of Arabidopsis in Spain and Germany under different density and rainfall conditions. In the milder German conditions, representatives of all accessions survived and reproduced, but in the harsher Spanish conditions only 193 of the 517 accessions survived. They then examined how natural selection affected allele frequencies. Interestingly, many of the alleles that were under positive selection in the harsh conditions are under negative selection in the mild conditions (a phenomenon known as antagonistic pleiotropy). Starting with their data and the data from similar experiments, the authors built genome-wide environment selection (GWES) models to predict natural selection at various sites, essentially simulating field experiments. The models indicate that at the edges of Arabidopsis’ range, populations are at evolutionary risk. The authors point out that this proof-of-concept experiment can be adapted to design conservation strategies for at-risk species. (Summary by Mary Williams) Nature 10.1038/s41586-019-1520-9