A rich evolutionary history of transposable element families in the maize genome (bioRxiv)
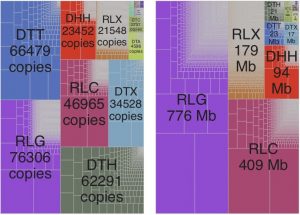 Transposable elements (TEs) make up nearly ~85% of the maize genome. Stitzer et al. report a comprehensive analysis of diverse TE families in maize based on the improved annotation of the updated B73 genome assembly. The authors found all of the known plant TE superfamilies in the maize genome, with Long Terminal Repeats (LTRs) occupying most of the genomic space, and Terminal Inverted Repeats (TIRs) accounting for the highest number of copies. In general, older TE family members coded for proteins whereas younger TE members were noncoding. As expected, most TE families exhibited high levels of DNA methylation, which limit their expression and activity. But a subset of TE families experienced variable levels and patterns of methylation. The authors attributed such variation as adaptation to the maize genomic environment each new TE experiences. These TE family annotations, and the associated analysis scripts can be further explored at https://mcstitzer.shinyapps.io/maize_te_families/, and https://github.com/mcstitzer/maize_genomic_ecosystem respectively. (Summary by Saima Shahid) bioRxiv 10.1101/559922.
Transposable elements (TEs) make up nearly ~85% of the maize genome. Stitzer et al. report a comprehensive analysis of diverse TE families in maize based on the improved annotation of the updated B73 genome assembly. The authors found all of the known plant TE superfamilies in the maize genome, with Long Terminal Repeats (LTRs) occupying most of the genomic space, and Terminal Inverted Repeats (TIRs) accounting for the highest number of copies. In general, older TE family members coded for proteins whereas younger TE members were noncoding. As expected, most TE families exhibited high levels of DNA methylation, which limit their expression and activity. But a subset of TE families experienced variable levels and patterns of methylation. The authors attributed such variation as adaptation to the maize genomic environment each new TE experiences. These TE family annotations, and the associated analysis scripts can be further explored at https://mcstitzer.shinyapps.io/maize_te_families/, and https://github.com/mcstitzer/maize_genomic_ecosystem respectively. (Summary by Saima Shahid) bioRxiv 10.1101/559922.