Plant Science Research Weekly: March 13
Opinion: We aren’t good at picking candidate genes, and it’s slowing us down
 Recent advances have facilitated the generation of huge phenotypic datasets from plant populations. However, the means to inexpensively organise such datasets to unequivocally determine causal genes has evaded researchers. Here, Baxter discusses how human bias when selecting candidate genes is compromising research efficiency. When utilizing breeding populations for association mapping, a genomic region correlated to a trait is often identified. Subsequently, genes localized in the region are compared to databases to reduce the number of candidates into a manageable list. This process requires human judgement when selecting what databases to use and which genes to follow up on, introducing bias. For example, we tend to focus on genes previously documented, limiting progress in characterising genes. The first step towards overcoming such biases is to recognise their existence and consciously work against them. Next, the author suggests potential solutions to shorten candidate lists without the need for human judgement, such as employing transcriptomics, proteomics, epigenetics and phylogenetics. (Summary by Caroline Dowling) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.01.006
Recent advances have facilitated the generation of huge phenotypic datasets from plant populations. However, the means to inexpensively organise such datasets to unequivocally determine causal genes has evaded researchers. Here, Baxter discusses how human bias when selecting candidate genes is compromising research efficiency. When utilizing breeding populations for association mapping, a genomic region correlated to a trait is often identified. Subsequently, genes localized in the region are compared to databases to reduce the number of candidates into a manageable list. This process requires human judgement when selecting what databases to use and which genes to follow up on, introducing bias. For example, we tend to focus on genes previously documented, limiting progress in characterising genes. The first step towards overcoming such biases is to recognise their existence and consciously work against them. Next, the author suggests potential solutions to shorten candidate lists without the need for human judgement, such as employing transcriptomics, proteomics, epigenetics and phylogenetics. (Summary by Caroline Dowling) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.01.006
Update: Twenty years of progress in physiological and biochemical investigation of RALF peptides
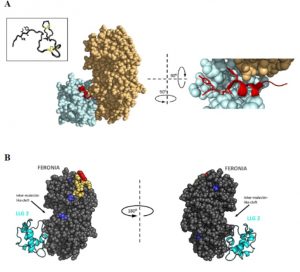 RALFs (Rapid Alkalinzation Factors) were one of the first types of signaling peptides identified in plants, and as their name suggests they were shown to induce an increase in the pH (i.e., alkalinization) of culture medium, through inducing phosphorylation of a plasma-membrane proton pump. There are more than 30 RALF-encoding genes in Arabidopsis, and also evidence that RALF-like sequences are present in some phytopathogenic fungi and bacteria. Blackburn et al. review the history of research into these important peptide signals, their receptors, and their biological functions. Key questions include how do the different peptides interact with closely related receptors, and what determines the specificity of these interactions and downstream functions. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.01310
RALFs (Rapid Alkalinzation Factors) were one of the first types of signaling peptides identified in plants, and as their name suggests they were shown to induce an increase in the pH (i.e., alkalinization) of culture medium, through inducing phosphorylation of a plasma-membrane proton pump. There are more than 30 RALF-encoding genes in Arabidopsis, and also evidence that RALF-like sequences are present in some phytopathogenic fungi and bacteria. Blackburn et al. review the history of research into these important peptide signals, their receptors, and their biological functions. Key questions include how do the different peptides interact with closely related receptors, and what determines the specificity of these interactions and downstream functions. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.01310
Review: Pathways to sex determination in plants: How many roads lead to Rome?
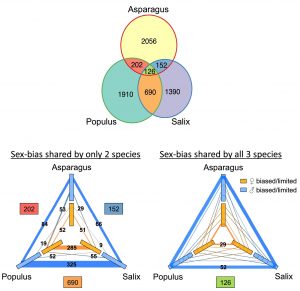 Although most angiosperms produce both male and female gametes, in some species an individual is either male or female, a property known as dioecy (literally, two houses). Dioecy exists in ~6% of angiosperms and is hypothesized to have evolved several times independently. While much research has been conducted to comprehend the molecular basis of sex determination in diverse species, insights into the commonalities and discrepancies between independent pathways are lacking. Feng et al. review our current understanding of sex determination, and hypothesize the existence of five models for the evolution of independent pathways. Furthermore, the authors discuss fifteen sex determination genes that have been detected in eight species, each representing distinct mechanisms to hinder gamete development. However, further evaluation revealed that six genes have roles in phytohormone (ethylene and cytokinin) regulation. Additionally, the authors re-analyzed published transcriptomes of three dioecious species with varying relatedness and sex chromosome structures (Asparagus an XY monocot, Salix and Populus are ZW and XY eudicots respectively). By investigating sex-biased gene expression, 126 orthogroups were found in all three species, and thus are candidates for conserved elements of sex regulation pathways throughout angiosperms. (Summary by Caroline Dowling) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.01.004
Although most angiosperms produce both male and female gametes, in some species an individual is either male or female, a property known as dioecy (literally, two houses). Dioecy exists in ~6% of angiosperms and is hypothesized to have evolved several times independently. While much research has been conducted to comprehend the molecular basis of sex determination in diverse species, insights into the commonalities and discrepancies between independent pathways are lacking. Feng et al. review our current understanding of sex determination, and hypothesize the existence of five models for the evolution of independent pathways. Furthermore, the authors discuss fifteen sex determination genes that have been detected in eight species, each representing distinct mechanisms to hinder gamete development. However, further evaluation revealed that six genes have roles in phytohormone (ethylene and cytokinin) regulation. Additionally, the authors re-analyzed published transcriptomes of three dioecious species with varying relatedness and sex chromosome structures (Asparagus an XY monocot, Salix and Populus are ZW and XY eudicots respectively). By investigating sex-biased gene expression, 126 orthogroups were found in all three species, and thus are candidates for conserved elements of sex regulation pathways throughout angiosperms. (Summary by Caroline Dowling) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.01.004
Opinion. Anthropogenic seed dispersal: Rethinking the origins of plant domestication
 Studies regarding the development of agriculture have started to integrate a plant evolutionary perspective. In this context, Spengler explores how changes in seed dispersal allowed plant domestication during the first half of the Holocene (e.g., more than 5000 years ago). He starts by redefining domestication as “the evolution of new traits to support a mutualistic relationship with humans“. The paper outlines four pathways that explain the changes in different crops. The first explains how robust reproductive structures allowed humans to domesticate mechanical-dispersed species, such as cereals and legumes. The second and third pathway present how humans became dispersers of large-fruited trees and herbs and how this impacted fruit size. Finally, the fourth pathway explores how ruminant-dispersed grasses increase seed size and loose dormancy in response to cultivation. With his paper, Spengler highlights that domestication is “part of the evolutionary arms race” and that embracing it like that would benefit future studies regarding agriculture’s origins. (Summary by Carlos A. Ordóñez-Parra) Trends Plant Sci. 10.1016/j.tplants.2020.01.00
Studies regarding the development of agriculture have started to integrate a plant evolutionary perspective. In this context, Spengler explores how changes in seed dispersal allowed plant domestication during the first half of the Holocene (e.g., more than 5000 years ago). He starts by redefining domestication as “the evolution of new traits to support a mutualistic relationship with humans“. The paper outlines four pathways that explain the changes in different crops. The first explains how robust reproductive structures allowed humans to domesticate mechanical-dispersed species, such as cereals and legumes. The second and third pathway present how humans became dispersers of large-fruited trees and herbs and how this impacted fruit size. Finally, the fourth pathway explores how ruminant-dispersed grasses increase seed size and loose dormancy in response to cultivation. With his paper, Spengler highlights that domestication is “part of the evolutionary arms race” and that embracing it like that would benefit future studies regarding agriculture’s origins. (Summary by Carlos A. Ordóñez-Parra) Trends Plant Sci. 10.1016/j.tplants.2020.01.00
Phenome analysis in multiple combinations of G-protein alpha and beta in Arabidopsis
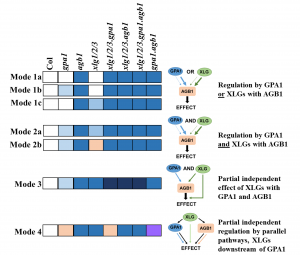 Heterotrimeric G-proteins are implicated in a large number of signalling pathways. As their name indicates, they are composed of three different subunits, Gα, Gβ, and Gγ. The Arabidopsis genome has genes encoding four Gα, one Gβ and two Gγ protein subunits, as compared to the multiple members that occur in human G-protein subunit families (23 Gα, 5 Gβ, 12 Gγ). Choudhury et al. generated different combinations of higher order mutants in the four Gα (including single Gα, GPA1 and three extra-large Gα, XLG1/2/3) and single Gβ (AGB1) subunits and compared these combinations of quadruple and quintuple mutants to the representative single, double and triple mutants. The authors summarise that G-protein subunits affect the plant phenotypes in a subunit-dependent manner and propose several mechanistic modes (Mode 1 to 4) to explain the complexity of the phenome network in G-protein signalling. This phenome analysis demonstrates the presence of various interactions between the Gα and the sole Gβ could influence specific pathways and contribute to the plant adaptation to different environmental changes. It would be interesting to do a similar study in a plant such as soybean that has multiple Gβ subunit. (Summary by Min May Wong) Plant Journal 10.1111/tpj.14714
Heterotrimeric G-proteins are implicated in a large number of signalling pathways. As their name indicates, they are composed of three different subunits, Gα, Gβ, and Gγ. The Arabidopsis genome has genes encoding four Gα, one Gβ and two Gγ protein subunits, as compared to the multiple members that occur in human G-protein subunit families (23 Gα, 5 Gβ, 12 Gγ). Choudhury et al. generated different combinations of higher order mutants in the four Gα (including single Gα, GPA1 and three extra-large Gα, XLG1/2/3) and single Gβ (AGB1) subunits and compared these combinations of quadruple and quintuple mutants to the representative single, double and triple mutants. The authors summarise that G-protein subunits affect the plant phenotypes in a subunit-dependent manner and propose several mechanistic modes (Mode 1 to 4) to explain the complexity of the phenome network in G-protein signalling. This phenome analysis demonstrates the presence of various interactions between the Gα and the sole Gβ could influence specific pathways and contribute to the plant adaptation to different environmental changes. It would be interesting to do a similar study in a plant such as soybean that has multiple Gβ subunit. (Summary by Min May Wong) Plant Journal 10.1111/tpj.14714
Transcription factor NF‐YB21 positively regulate the root growth in Populus
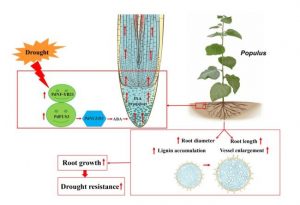 Nuclear factor Y (NF-Y) proteins are heterotrimeric transcription factors made up of A, B and C subunits that exist in higher eukaryotes. Previous work has implicated NF-Ys in root growth. Recently, Zhou et al. isolated a root-specific NF-Y family transcription factor in Populus designated as PdNF-YB21. Overexpression or knockout of PdNF-YB21 results in enhanced or reduced root growth, xylem lignification, and drought resistance respectively. Moreover, PdNF-YB21 interacts with the transcription factor PdFUS3, which activates the promoter of the key ABA synthesis gene PdNCED3. ABA promotes IAA transport in root tips, which ultimately increases root growth and drought resistance. This study revealed a functional module, NF‐YB21‐FUS3‐NCED3, in poplar root growth in response to water-deficit, and also shows how ABA synthesis in Populus influences auxin transport.(Summary by Nanxun Qin) New Phytol. 10.1111/nph.16524
Nuclear factor Y (NF-Y) proteins are heterotrimeric transcription factors made up of A, B and C subunits that exist in higher eukaryotes. Previous work has implicated NF-Ys in root growth. Recently, Zhou et al. isolated a root-specific NF-Y family transcription factor in Populus designated as PdNF-YB21. Overexpression or knockout of PdNF-YB21 results in enhanced or reduced root growth, xylem lignification, and drought resistance respectively. Moreover, PdNF-YB21 interacts with the transcription factor PdFUS3, which activates the promoter of the key ABA synthesis gene PdNCED3. ABA promotes IAA transport in root tips, which ultimately increases root growth and drought resistance. This study revealed a functional module, NF‐YB21‐FUS3‐NCED3, in poplar root growth in response to water-deficit, and also shows how ABA synthesis in Populus influences auxin transport.(Summary by Nanxun Qin) New Phytol. 10.1111/nph.16524
In Arabidopsis, low blue light enhances phototropism by releasing cryptochrome 1-mediated inhibition of PIF4 expression

A plant’s light environment is complex and variable, but through different photoreceptors the plant can perceive its environment and grow appropriately. For example, plants respond differently in the presence of dense vegetation (leading to a low red to far-red light ratio due to far-red light being reflected off nearby leaves) versus canopy shade (low levels of red and blue light as well as low red to far-red ratio). But in canopy shade, unfiltered light can pass through the gaps, and in this case, plants tend to reorient their stem towards the available light in what is known as phototropism. In this paper, Boccaccini et al. determined how canopy shade promotes phototropism. Experimental setups mimicking natural sunlight, canopy shade, low blue light, or low red to far-red ratio showed increased hypocotyl reorientation towards low blue light and canopy shade. Further tests with loss of function mutant lines, cry1 (Cryptochrome 1), phot1 (Phototropin 1), nph3 (NPH1-Photoreceptor interacting protein), pif4pif5pif6 (Phytochrome interacting factors) for their hypocotyl reorientation under low blue light was performed. Overall the authors have shown that phototropic enhancement during low-blue light canopy shade requires PHOT1 to perceive the blue light gradient which causes a reduction in CRY1 activity. This enhances PIF4 abundance leading to the modification of auxin related genes which promotes hypocotyl reorientation towards the unfiltered light during canopy shade. (Summary by Sunita Pathak) bioRxiv 10.1101/2020.02.28.969725
Heat tolerance regulated by an ancient jasmonate signaling pathway
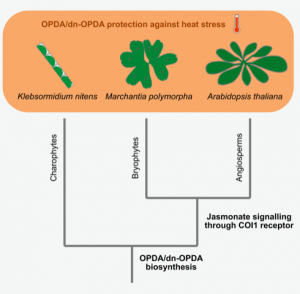 Jasmonate responses are regulated not only by the well-studied pair of the bioactive hormone jasmonoyl-isoleucine (JA-Ile) and the receptor COI1, but also the cyclopentenone OPDA and the JA-Ile precursor dn-OPDA can activate jasmonate signaling. However, the OPDA/dn-OPDA signaling and their physiological roles are not fully understood. To dissect the COI1-independent OPDA/dn-OPDA responses, Monte, Kneeshaw, et al. capitalized on recently developed resources: the liverwort Marchantia polymorpha coi1 mutants and the eudicot Arabidopsis thaliana opr3-3 coi1 double mutants, both of which produce OPDA/dn-OPDA but are unable to perceive JA-Ile, as well as the charophyte alga Klebsormidium, which does not have COI1 but can produce OPDA/dn-OPDA. The authors found that OPDA/dn-OPDA can enhance heat stress tolerance in both M. polymorpha and Klebsormidium in a COI1-independent manner. Transcriptome analysis showed that COI1-independent OPDA responses, including heat stress responses, were conserved among M. polymorpha, Klebsormidium, and A. thaliana. An author noted on Twitter “Once upon a time our growth chamber broke, the temperature increased, and only the Marchantia plants grown on OPDA survived.” This serendipitous finding enriches our understanding of an ancestral role of jasmonate signaling that may have been key to land plant colonization. (Summary by Tatsuya Nobori) Current Biology 10.1016/j.cub.2020.01.023
Jasmonate responses are regulated not only by the well-studied pair of the bioactive hormone jasmonoyl-isoleucine (JA-Ile) and the receptor COI1, but also the cyclopentenone OPDA and the JA-Ile precursor dn-OPDA can activate jasmonate signaling. However, the OPDA/dn-OPDA signaling and their physiological roles are not fully understood. To dissect the COI1-independent OPDA/dn-OPDA responses, Monte, Kneeshaw, et al. capitalized on recently developed resources: the liverwort Marchantia polymorpha coi1 mutants and the eudicot Arabidopsis thaliana opr3-3 coi1 double mutants, both of which produce OPDA/dn-OPDA but are unable to perceive JA-Ile, as well as the charophyte alga Klebsormidium, which does not have COI1 but can produce OPDA/dn-OPDA. The authors found that OPDA/dn-OPDA can enhance heat stress tolerance in both M. polymorpha and Klebsormidium in a COI1-independent manner. Transcriptome analysis showed that COI1-independent OPDA responses, including heat stress responses, were conserved among M. polymorpha, Klebsormidium, and A. thaliana. An author noted on Twitter “Once upon a time our growth chamber broke, the temperature increased, and only the Marchantia plants grown on OPDA survived.” This serendipitous finding enriches our understanding of an ancestral role of jasmonate signaling that may have been key to land plant colonization. (Summary by Tatsuya Nobori) Current Biology 10.1016/j.cub.2020.01.023
Allelic mutations in the Ripening-inhibitor (RIN) locus generate extensive variation in tomato ripening ($)
 Ripened fruits attract animals to eat and disperse seeds, allowing propagation. Slowing down the fruit ripening process is often used commercially to decrease damage during transport and extend shelf life. Molecular (increased pigment, aroma, and flavor) and physiological (softened flesh) changes of fruit ripening are highly synchronized, and RIPENING INHIBITOR (RIN) is one of the transcription factors regulating the process. Ito et al. used CRISPR/Cas9 to systematically generate and characterize rin mutants, including a previously-identified non-ripening mutant (rin), knockout mutants (KO), and mutants lacking the activator domain (rinG2). KO showed exacerbated flesh softening whereas rinG2 flesh remained unchanged along with high levels of carotenoid accumulation. The phenotypic disparities between rin mutants prompted the hypothesis that “RIN is not only a ripening activator, but also acts as a repressor of over-softening.” The study proposes a dual regulatory model involving RIN and ethylene in regulating fruit ripening-related genes, and provides an applicable strategy to tune activities of transcription factors. Further, the KO and rinG2 mutations could be adapted for enhances in fruit production. (Summary by Yun-Ting Kao) Plant Physiol. 10.1104/pp.20.00020
Ripened fruits attract animals to eat and disperse seeds, allowing propagation. Slowing down the fruit ripening process is often used commercially to decrease damage during transport and extend shelf life. Molecular (increased pigment, aroma, and flavor) and physiological (softened flesh) changes of fruit ripening are highly synchronized, and RIPENING INHIBITOR (RIN) is one of the transcription factors regulating the process. Ito et al. used CRISPR/Cas9 to systematically generate and characterize rin mutants, including a previously-identified non-ripening mutant (rin), knockout mutants (KO), and mutants lacking the activator domain (rinG2). KO showed exacerbated flesh softening whereas rinG2 flesh remained unchanged along with high levels of carotenoid accumulation. The phenotypic disparities between rin mutants prompted the hypothesis that “RIN is not only a ripening activator, but also acts as a repressor of over-softening.” The study proposes a dual regulatory model involving RIN and ethylene in regulating fruit ripening-related genes, and provides an applicable strategy to tune activities of transcription factors. Further, the KO and rinG2 mutations could be adapted for enhances in fruit production. (Summary by Yun-Ting Kao) Plant Physiol. 10.1104/pp.20.00020
Disturbance of floral color pattern by activation of an endogenous virus in aged petunia plants
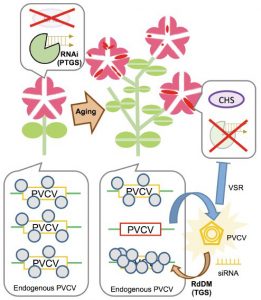 The red color of petunia petals is due to the presence of anthocyanin pigments, produced in part through the action of the chalcone synthase enzyme. In the “Red Star”’ variety, expression of the CHS-A gene encoding chalcone synthase is silenced in cells around the midvein through post-transcriptional gene silencing (PTGS). Plant viruses can inhibit naturally occurring PTGS by viral suppressors of RNA silencing (VSRs) to protect themselves. In this work, Kuriyama et al. characterized the activation process of petunia vein clearing virus (PVCV), an endogenous pararetrovirus, and the effect of its activation on the bicolor pattern of star-type petunia; they observed that in aged plants, anthocyanin accumulates in the normally white petal regions. The authors identified a decrease in DNA methylation level in the promoter region of proviral PVCV in aged petunia plants, supporting the interpretation that the virus is activated. PVCV transcripts and episomal DNA accumulation were found in regions of blotched petals, where the expression of CHS-A was also restored. This indicates PTGS suppression by VSRs activity; in other words, the gene, normally silenced by PTGS, is switched on because VSR suppresses PTGS. The appearance of blotched flowers show that in aged plants, PVCV have won the tug-of-war between a plant host and a pathogen. (Summary by Elisandra Pradella) Plant Journal 10.1111/tpj.14728
The red color of petunia petals is due to the presence of anthocyanin pigments, produced in part through the action of the chalcone synthase enzyme. In the “Red Star”’ variety, expression of the CHS-A gene encoding chalcone synthase is silenced in cells around the midvein through post-transcriptional gene silencing (PTGS). Plant viruses can inhibit naturally occurring PTGS by viral suppressors of RNA silencing (VSRs) to protect themselves. In this work, Kuriyama et al. characterized the activation process of petunia vein clearing virus (PVCV), an endogenous pararetrovirus, and the effect of its activation on the bicolor pattern of star-type petunia; they observed that in aged plants, anthocyanin accumulates in the normally white petal regions. The authors identified a decrease in DNA methylation level in the promoter region of proviral PVCV in aged petunia plants, supporting the interpretation that the virus is activated. PVCV transcripts and episomal DNA accumulation were found in regions of blotched petals, where the expression of CHS-A was also restored. This indicates PTGS suppression by VSRs activity; in other words, the gene, normally silenced by PTGS, is switched on because VSR suppresses PTGS. The appearance of blotched flowers show that in aged plants, PVCV have won the tug-of-war between a plant host and a pathogen. (Summary by Elisandra Pradella) Plant Journal 10.1111/tpj.14728
An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages ($)
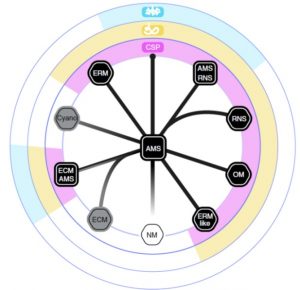 It’s widely thought that plants acquired the ability to live on land thanks to a little help from their friends, specifically arbuscular mycorrhizal fungi. Even now, most land plants form mutually beneficial associations with fungi or bacteria, and these often involve the plant cells acting as hosts for their intracellular symbionts. Some plants however have lost this ability, in what is termed mutualism abandonment. Radhakrishnan et al. examined more than 270 transcriptomes and 100 genomes spanning the plant kingdom to evaluate the presence of a core set of genes previously identified as required for arbuscular mycorrhizal symbiosis. Three of these are present in all plants that form intracellular symbioses, including those with a distinct origin such as ericoid myorrhizae, indicating “conservation in their evolution across 450 million years of plant diversification.” However, in species that have lost the ability to form intracellular symbioses, such as ectomycorrhizal plants in which the symbiotic fungus doesn’t enter the plant cells, these conserved core genes have been lost, indicating the involvement of a distinct molecular machinery. (Summary by Mary Williams) Nature Plants 10.1038/s41477-020-0613-7
It’s widely thought that plants acquired the ability to live on land thanks to a little help from their friends, specifically arbuscular mycorrhizal fungi. Even now, most land plants form mutually beneficial associations with fungi or bacteria, and these often involve the plant cells acting as hosts for their intracellular symbionts. Some plants however have lost this ability, in what is termed mutualism abandonment. Radhakrishnan et al. examined more than 270 transcriptomes and 100 genomes spanning the plant kingdom to evaluate the presence of a core set of genes previously identified as required for arbuscular mycorrhizal symbiosis. Three of these are present in all plants that form intracellular symbioses, including those with a distinct origin such as ericoid myorrhizae, indicating “conservation in their evolution across 450 million years of plant diversification.” However, in species that have lost the ability to form intracellular symbioses, such as ectomycorrhizal plants in which the symbiotic fungus doesn’t enter the plant cells, these conserved core genes have been lost, indicating the involvement of a distinct molecular machinery. (Summary by Mary Williams) Nature Plants 10.1038/s41477-020-0613-7