Plant Science Research Weekly: July 7, 2023
Review. CRISPR/Cas-mediated plant genome editing: Outstanding challenges a decade after implementation
 I’ll be honest, I was surprised to see “a decade after implementation” in this title, but indeed, the first publication describing CRISPR/Cas in plants was in 2013. We’ve learned a lot in the past 10 years and the technology provides many opportunities, but challenges remain, both of which are highlighted in this excellent review by Cardi et al. One of the most impressive developments has been that of advanced CRISPR/Cas variants capable of modifying DNA in various ways such as prime editing and introduction of deletions. An ongoing challenge remains in how to introduce the editing enzymes and guide RNAs into plant cells, and the review does an excellent job of summarizing several options. Regeneration of the transformed or edited plants through tissue culture is yet another challenge, although the use of developmental regulators (BABY BOOM, SHOOT MERISTEMLESS) can help. Various methods to detect the outcomes, including unintended consequences, are discussed, as well as several excellent and important applications of gene editing technologies in plants (Summary by Mary Williams @PlantTeaching) Trends Plant Sci 10.1016/j.tplants.2023.05.012
I’ll be honest, I was surprised to see “a decade after implementation” in this title, but indeed, the first publication describing CRISPR/Cas in plants was in 2013. We’ve learned a lot in the past 10 years and the technology provides many opportunities, but challenges remain, both of which are highlighted in this excellent review by Cardi et al. One of the most impressive developments has been that of advanced CRISPR/Cas variants capable of modifying DNA in various ways such as prime editing and introduction of deletions. An ongoing challenge remains in how to introduce the editing enzymes and guide RNAs into plant cells, and the review does an excellent job of summarizing several options. Regeneration of the transformed or edited plants through tissue culture is yet another challenge, although the use of developmental regulators (BABY BOOM, SHOOT MERISTEMLESS) can help. Various methods to detect the outcomes, including unintended consequences, are discussed, as well as several excellent and important applications of gene editing technologies in plants (Summary by Mary Williams @PlantTeaching) Trends Plant Sci 10.1016/j.tplants.2023.05.012
Plant Physiology Focus Issue: Fruit Crops
 July brings delicious fruit harvests in the Northern Hemisphere, and a very special focus issue of Plant Physiology. I particularly like this issue because of the wide variety of species covered, starting with apple, banana, blueberry, cherry, citrus, and so on. It’s a nice departure from our usual “diet” of wheat, rice, and Arabidopsis. The update and research articles encompass the range of topics that affect fruit production and quality: drought responses, breeding technologies, biotic stresses, ripening/softening, and flavor molecules and phytonutrients. The editors have invited six speakers whose work appears in this focus issue to present their work in a pair of webinars to be held on July 12 (https://plantae.org/plant-physiology-focus-issue-webinar-fruit-crops-1/) and July 27. (Summary by Mary Williams @PlantTeaching) Plant Physiology https://academic.oup.com/plphys/issue/192/3
July brings delicious fruit harvests in the Northern Hemisphere, and a very special focus issue of Plant Physiology. I particularly like this issue because of the wide variety of species covered, starting with apple, banana, blueberry, cherry, citrus, and so on. It’s a nice departure from our usual “diet” of wheat, rice, and Arabidopsis. The update and research articles encompass the range of topics that affect fruit production and quality: drought responses, breeding technologies, biotic stresses, ripening/softening, and flavor molecules and phytonutrients. The editors have invited six speakers whose work appears in this focus issue to present their work in a pair of webinars to be held on July 12 (https://plantae.org/plant-physiology-focus-issue-webinar-fruit-crops-1/) and July 27. (Summary by Mary Williams @PlantTeaching) Plant Physiology https://academic.oup.com/plphys/issue/192/3
Brassinosteroid coordinates cell layer interactions via cell wall and tissue mechanics
 Organismal growth requires extensive coordination between cells and tissues with different identities, and this is particularly important for plants with their rigid cell walls and lack of cell motility. A key mechanism for tissue coordination involving brassinosteroids has been identified by Kelly-Bellow et al. They started by looking at a dwarf mutant (isolated from an EMS-mutagenesis screen) of Utricularia gibba, an aquatic carnivorous angiosperm also known as humped bladderwort. The dwarf plants showed decreased epidermal cell elongation and the airspace-containing internal tissues were twisted. Modeling studies suggest that the phenotype is caused by inhibited growth in the epidermis. The affected DWARF gene encodes an enzyme involved in brassinosteroid biosynthesis, and the authors propose that brassinosteroids reduce mechanical epidermal constraint. Although a similar effect isn’t seen in brassinosteroid-deficient Arabidopsis plants, the Arabidopsis stems lack air spaces, so the internal tissues are held together more rigidly than in the floating aquatic plant stems. Therefore, the authors crossed a mutant that affects cell adhesion (quasimodo) with brassinosteroid deficient lines, leading to dramatic cracks in the epidermis as beautifully illustrated on the cover of Science. The authors propose that brassinosteroids alleviate growth constraints through cell wall modifications, and conclude that “Gene activity may therefore have coordinated effects on tissue development not only via molecular signaling but also via mechanics.” (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.adf0752
Organismal growth requires extensive coordination between cells and tissues with different identities, and this is particularly important for plants with their rigid cell walls and lack of cell motility. A key mechanism for tissue coordination involving brassinosteroids has been identified by Kelly-Bellow et al. They started by looking at a dwarf mutant (isolated from an EMS-mutagenesis screen) of Utricularia gibba, an aquatic carnivorous angiosperm also known as humped bladderwort. The dwarf plants showed decreased epidermal cell elongation and the airspace-containing internal tissues were twisted. Modeling studies suggest that the phenotype is caused by inhibited growth in the epidermis. The affected DWARF gene encodes an enzyme involved in brassinosteroid biosynthesis, and the authors propose that brassinosteroids reduce mechanical epidermal constraint. Although a similar effect isn’t seen in brassinosteroid-deficient Arabidopsis plants, the Arabidopsis stems lack air spaces, so the internal tissues are held together more rigidly than in the floating aquatic plant stems. Therefore, the authors crossed a mutant that affects cell adhesion (quasimodo) with brassinosteroid deficient lines, leading to dramatic cracks in the epidermis as beautifully illustrated on the cover of Science. The authors propose that brassinosteroids alleviate growth constraints through cell wall modifications, and conclude that “Gene activity may therefore have coordinated effects on tissue development not only via molecular signaling but also via mechanics.” (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.adf0752
Light and sucrose signaling converge at TOR kinase to control plant development
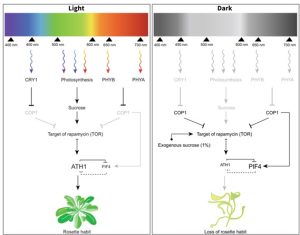 Different photoreceptors (mainly phytochromes and cryptochromes) perceive light, which acts as a signal for controlling plant growth and development. Similarly, light is absorbed by chlorophylls (and carotenoids) for generating energy via photosynthesis. In search of a link between these two light-mediated processes, Hajibehzad et al. explored a gene, ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) induced by both light and sugars, and controlling the rosette habit in Arabidopsis. Similar morphology (lack of rosette habit) in ath1 and photoreceptor (phy, cry) mutants revealed a double negative feedback regulation between ATH1 and PHYTOCHROME INTERACTING FACTORS (PIFs) in the shoot apical meristem (SAM) which can possibly act as a switch during the vegetative to floral transition phase. High sugar availability to the SAM can overcome the light requirement for ATH1 expression. Increased expression of ATH1 and a related increase in the inhibition of vegetative internode elongation occurred in response to rising sucrose concentrations. Light and sucrose can act as signals as well as energy sources. Through strategically defined experiments, the group identified that the rosette habit was controlled by the signaling property of light and the energy property of both light and sucrose. TARGET OF RAPAMYCIN (TOR) kinase, a central component in energy sensing, must be activated for ATH1 to be induced at the SAM by both metabolic and light signals. Thus, light and sucrose signaling pathways converge at TOR kinase to control ATH1 expression and subsequent rosette growth habit in Arabidopsis thaliana. (Summary by Rajarshi Sanyal, @rajarshi_sanyal) New Phytologist 10.1111/nph.19014)
Different photoreceptors (mainly phytochromes and cryptochromes) perceive light, which acts as a signal for controlling plant growth and development. Similarly, light is absorbed by chlorophylls (and carotenoids) for generating energy via photosynthesis. In search of a link between these two light-mediated processes, Hajibehzad et al. explored a gene, ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) induced by both light and sugars, and controlling the rosette habit in Arabidopsis. Similar morphology (lack of rosette habit) in ath1 and photoreceptor (phy, cry) mutants revealed a double negative feedback regulation between ATH1 and PHYTOCHROME INTERACTING FACTORS (PIFs) in the shoot apical meristem (SAM) which can possibly act as a switch during the vegetative to floral transition phase. High sugar availability to the SAM can overcome the light requirement for ATH1 expression. Increased expression of ATH1 and a related increase in the inhibition of vegetative internode elongation occurred in response to rising sucrose concentrations. Light and sucrose can act as signals as well as energy sources. Through strategically defined experiments, the group identified that the rosette habit was controlled by the signaling property of light and the energy property of both light and sucrose. TARGET OF RAPAMYCIN (TOR) kinase, a central component in energy sensing, must be activated for ATH1 to be induced at the SAM by both metabolic and light signals. Thus, light and sucrose signaling pathways converge at TOR kinase to control ATH1 expression and subsequent rosette growth habit in Arabidopsis thaliana. (Summary by Rajarshi Sanyal, @rajarshi_sanyal) New Phytologist 10.1111/nph.19014)
RHO GTPase of plants regulates polarized cell growth and cell division orientation during morphogenesis
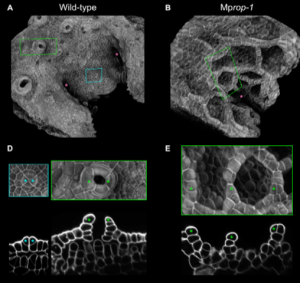 Precise spatial control of cell division and cell growth is necessary to produce the specific cellular organizations demanded by the complex tissues and organs of morphologically complex organisms One of many factors that guide cell division/growth is cell polarity, of which RHO GTPase-type proteins have emerged as conserved eukaryotic regulators. RHO of Plants (ROP) form a plant-specific clade of RHO GTPases required for cell polarization in different species and developmental contexts. ROP proteins achieve polarization by localizing to specific cellular domains, however, although ROP function has been investigated in different cellular contexts no clear phenotypes have been observed at the level of plant tissues/organs even in higher order rop mutants, leading to the question: do ROP proteins contribute to the formation of complex tissues and/or organs? Seeking an answer to this question Mulvey and Dolan took advantage of the liverwort (Marchantia polymorpha) low genetic redundancy, which encodes a single ROP gene (MpROP). Mprop mutants display an array of tissular and morphological phenotypes resulting in defective air chambers, gemma cups and gemmae, consistent with the authors’ hypothesis that MpROP participates in organogenesis and tissue development. Detailed morphometric analyses of Mprop and fine imaging of translational reporter and overexpression lines showed that MpROP localizes at sites of polarized growth and gradually accumulates at the expanding cell plate as the cell prepares to divide. Consistent with the observed subcellular localization, Mprop mutants display misoriented cell divisions and loss of polarized growth, whereas MpROP overexpression also produces misoriented cell divisions. Combining reverse genetics with elegant imaging the authors were able to shed light on a likely conserved role of ROP protein in polarized growth and division orientation regulation to guide proper development of complex tissues and organs in land plants, a function that has been likely obscured by genetic redundancy in other models. Summary by Jesús León @jesussaur) Curr. Biol. 10.1016/j.cub.2023.06.015
Precise spatial control of cell division and cell growth is necessary to produce the specific cellular organizations demanded by the complex tissues and organs of morphologically complex organisms One of many factors that guide cell division/growth is cell polarity, of which RHO GTPase-type proteins have emerged as conserved eukaryotic regulators. RHO of Plants (ROP) form a plant-specific clade of RHO GTPases required for cell polarization in different species and developmental contexts. ROP proteins achieve polarization by localizing to specific cellular domains, however, although ROP function has been investigated in different cellular contexts no clear phenotypes have been observed at the level of plant tissues/organs even in higher order rop mutants, leading to the question: do ROP proteins contribute to the formation of complex tissues and/or organs? Seeking an answer to this question Mulvey and Dolan took advantage of the liverwort (Marchantia polymorpha) low genetic redundancy, which encodes a single ROP gene (MpROP). Mprop mutants display an array of tissular and morphological phenotypes resulting in defective air chambers, gemma cups and gemmae, consistent with the authors’ hypothesis that MpROP participates in organogenesis and tissue development. Detailed morphometric analyses of Mprop and fine imaging of translational reporter and overexpression lines showed that MpROP localizes at sites of polarized growth and gradually accumulates at the expanding cell plate as the cell prepares to divide. Consistent with the observed subcellular localization, Mprop mutants display misoriented cell divisions and loss of polarized growth, whereas MpROP overexpression also produces misoriented cell divisions. Combining reverse genetics with elegant imaging the authors were able to shed light on a likely conserved role of ROP protein in polarized growth and division orientation regulation to guide proper development of complex tissues and organs in land plants, a function that has been likely obscured by genetic redundancy in other models. Summary by Jesús León @jesussaur) Curr. Biol. 10.1016/j.cub.2023.06.015
Comparative phylotranscriptomics reveals ancestral and derived root nodule symbiosis programs
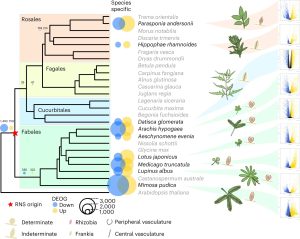 There are about ~17,500 plants species that can participate in nitrogen-fixing root nodule symbioses. The majority of these (~17,300) are in the order Fabales, which includes the legumes. The remainder fall into three orders (Rosales, Fagales, and Cucubitales), leading to the question of whether this ability in these outliers is derived from the same evolutionary event or is the result of convergent processes. A new study from Libourel and Keller et al. addresses this question, in part by obtaining the sequence of Mimosa pudica, part of the Mimosoid clade of legumes that did not have an available genomic sequence. The authors carried out thorough transcriptomic studies involving it and eight other nodulating species including three outside the Fabales. They identified several hundred orthogroups (sets of genes from multiple species descended from a single gene in the last common ancestor of that set of species) ancestrally upregulated during root nodule symbiosis. The main conclusion is that most of the steps involved in root nodule symbiosis were very likely present in the shared common ancestor, and that there have been very high rates of symbiosis loss from its descendants. There are many additional findings presented, such a discussion of the ancestral function of some of these genes, including among others lateral root development and arbuscular mycorrhizal symbiosis, and more recent evolutionary innovations that support the success or not of these symbioses. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-023-01441-w
There are about ~17,500 plants species that can participate in nitrogen-fixing root nodule symbioses. The majority of these (~17,300) are in the order Fabales, which includes the legumes. The remainder fall into three orders (Rosales, Fagales, and Cucubitales), leading to the question of whether this ability in these outliers is derived from the same evolutionary event or is the result of convergent processes. A new study from Libourel and Keller et al. addresses this question, in part by obtaining the sequence of Mimosa pudica, part of the Mimosoid clade of legumes that did not have an available genomic sequence. The authors carried out thorough transcriptomic studies involving it and eight other nodulating species including three outside the Fabales. They identified several hundred orthogroups (sets of genes from multiple species descended from a single gene in the last common ancestor of that set of species) ancestrally upregulated during root nodule symbiosis. The main conclusion is that most of the steps involved in root nodule symbiosis were very likely present in the shared common ancestor, and that there have been very high rates of symbiosis loss from its descendants. There are many additional findings presented, such a discussion of the ancestral function of some of these genes, including among others lateral root development and arbuscular mycorrhizal symbiosis, and more recent evolutionary innovations that support the success or not of these symbioses. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-023-01441-w
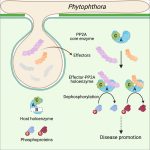
Pathogen protein modularity enables mimicry of a host phosphatase
 Some of the most fascinating discoveries in biology are found at the interface between hosts and pathogens, where each organism is endlessly fighting for its survival. Plant pathogens produce a wide array of effector proteins that promote their virulence, but we only understand how some of these effectors function. Here, Li et al. have uncovered how a family of effectors from the oomycete pathogen Phytophthora sojae promote disease in their hosts. Previous studies identified a large family of effector proteins with stretches of repeating units consisting of several L (Leu) residues followed by one W (Trp) and one Y (Tyr), known as (L)WY motifs. The authors found that some of these (L)WY-motif containing effectors interact with the core complex of the serine/threonine protein phosphatase 2A (PP2A) enzyme. Specifically, these effectors compete with the regulatory B domain, which specifies the dephosphorylation targets, for binding to the core complex. The presence of the effectors changes the specificity of the core phosphatase, leading to different sets of dephosphorylated proteins; different from in the absence of the effector, and different between different (L)WY-containing effectors. The authors point out that the modularity of these effector domains readily permits domain shuffling to generate new effectors conferring different specificity to the hybrid phosphastase complex, enhancing the evolvability of the effector repertoire. (Summary by Mary Williams @PlantTeaching) Cell 10.1016/j.cell.2023.05.049
Some of the most fascinating discoveries in biology are found at the interface between hosts and pathogens, where each organism is endlessly fighting for its survival. Plant pathogens produce a wide array of effector proteins that promote their virulence, but we only understand how some of these effectors function. Here, Li et al. have uncovered how a family of effectors from the oomycete pathogen Phytophthora sojae promote disease in their hosts. Previous studies identified a large family of effector proteins with stretches of repeating units consisting of several L (Leu) residues followed by one W (Trp) and one Y (Tyr), known as (L)WY motifs. The authors found that some of these (L)WY-motif containing effectors interact with the core complex of the serine/threonine protein phosphatase 2A (PP2A) enzyme. Specifically, these effectors compete with the regulatory B domain, which specifies the dephosphorylation targets, for binding to the core complex. The presence of the effectors changes the specificity of the core phosphatase, leading to different sets of dephosphorylated proteins; different from in the absence of the effector, and different between different (L)WY-containing effectors. The authors point out that the modularity of these effector domains readily permits domain shuffling to generate new effectors conferring different specificity to the hybrid phosphastase complex, enhancing the evolvability of the effector repertoire. (Summary by Mary Williams @PlantTeaching) Cell 10.1016/j.cell.2023.05.049