Plant Science Research Weekly: February 28
Review: Crop phenomics and high-throughput phenotyping
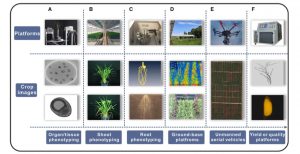 Crop phenomics has lagged behind crop genomics because traditional methods are time-consuming, expensive, invasive and subjective. Recently, high-throughput, automated, sensor and machine-vision methods have been developed, as reviewed by Yang et al. This review describes a large number of phenotyping platforms, including the early efforts, and compares their applications, strengths and weakenesses. (It’s interesting to see how rapidly these technologies have developed and become faster, better, and more reliable.) They also discuss various sensors and imaging methods (red-green-blue, thermal, hyperspectral), different ways to analyze roots (CT, MRI, PET), field-level sensing using towers or mobile carts, and remote sensing using drones. Post-harvest phenotyping and combined phenotyping-genotyping methods are also discussed. This is an excellent and accessible introduction into the exiting arena of crop phenomics and well worth a read. (Summary by Mary Williams) Mol. Plant 10.1016/j.molp.2020.01.008
Crop phenomics has lagged behind crop genomics because traditional methods are time-consuming, expensive, invasive and subjective. Recently, high-throughput, automated, sensor and machine-vision methods have been developed, as reviewed by Yang et al. This review describes a large number of phenotyping platforms, including the early efforts, and compares their applications, strengths and weakenesses. (It’s interesting to see how rapidly these technologies have developed and become faster, better, and more reliable.) They also discuss various sensors and imaging methods (red-green-blue, thermal, hyperspectral), different ways to analyze roots (CT, MRI, PET), field-level sensing using towers or mobile carts, and remote sensing using drones. Post-harvest phenotyping and combined phenotyping-genotyping methods are also discussed. This is an excellent and accessible introduction into the exiting arena of crop phenomics and well worth a read. (Summary by Mary Williams) Mol. Plant 10.1016/j.molp.2020.01.008
Review: Tropical trees as time capsules of anthropogenic activity
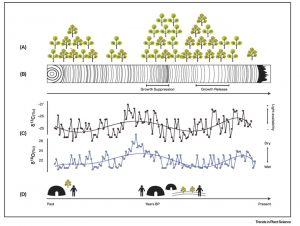 Trees will help us ensure our future, but they are also a valuable record of our past. This fascinating review article by Caetano-Andrade et al. describes how anthropologists are taking advantage of data recorded in trees to understand more about not only the atmospheric and geological events of the past, but also the human activities. A classic method for reading tree data is dendochrononlogy, or tree-ring data, in which the sizes of annual rings provides insights into the growing seasons; this method is strengthened when combined with radiocarbon data. These can shed insights into human activities including deforestation. Stable isotopes analysis (i.e., the difference in accumulation between light and heavy isotopes of water and carbon, δ18O and δ13C) gives insights into water and light availability, providing records of seasonal climate variations (e.g., El Niño events, droughts and monsoons), which can be causes or consequences of human activities. DNA analysis is also useful, and can for example indicate periods of rapid population bottlenecks perhaps as a consequence of logging, or provide evidence of domestication. The authors provide numerous examples for how trees have revealed stories of human activities, and encourage the protection of these “cultural heritage repositories.” (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2019.12.010
Trees will help us ensure our future, but they are also a valuable record of our past. This fascinating review article by Caetano-Andrade et al. describes how anthropologists are taking advantage of data recorded in trees to understand more about not only the atmospheric and geological events of the past, but also the human activities. A classic method for reading tree data is dendochrononlogy, or tree-ring data, in which the sizes of annual rings provides insights into the growing seasons; this method is strengthened when combined with radiocarbon data. These can shed insights into human activities including deforestation. Stable isotopes analysis (i.e., the difference in accumulation between light and heavy isotopes of water and carbon, δ18O and δ13C) gives insights into water and light availability, providing records of seasonal climate variations (e.g., El Niño events, droughts and monsoons), which can be causes or consequences of human activities. DNA analysis is also useful, and can for example indicate periods of rapid population bottlenecks perhaps as a consequence of logging, or provide evidence of domestication. The authors provide numerous examples for how trees have revealed stories of human activities, and encourage the protection of these “cultural heritage repositories.” (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2019.12.010
Metabolic labeling of RNAs uncovers hidden features and dynamics of the Arabidopsis transcriptome
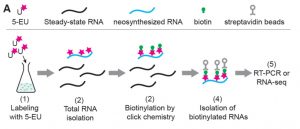 The ability to directly sequence RNAs (RNA-seq) has revolutionized our understanding of gene expression, but it can miss or underestimate short-lived RNAs. Several methods have been developed to identify newly-synthesized mRNAs to provide a snapshot of transcription as it happens. Szabo present Neu-seq, an in vivo pulse-labeling method using a non-toxic uridine analog (5-EU; 5-ethynyl uridine) followed by biotin purification that detects neosynthesized transcripts, short-lived transcripts, and processing intermediates (e.g., incompletely-spliced transcripts). The method can also be used to estimate rates of RNA decay. The authors address the benefits of this method as compared to other similar methods, for example the use of transcriptional inhibitors, and they also observe that the method is suitable for detection of plastid transcription. As one reviewer observed, “This method will likely have a large impact on the field of nuclear/organellar RNA metabolism, and the subject is very timely.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00214
The ability to directly sequence RNAs (RNA-seq) has revolutionized our understanding of gene expression, but it can miss or underestimate short-lived RNAs. Several methods have been developed to identify newly-synthesized mRNAs to provide a snapshot of transcription as it happens. Szabo present Neu-seq, an in vivo pulse-labeling method using a non-toxic uridine analog (5-EU; 5-ethynyl uridine) followed by biotin purification that detects neosynthesized transcripts, short-lived transcripts, and processing intermediates (e.g., incompletely-spliced transcripts). The method can also be used to estimate rates of RNA decay. The authors address the benefits of this method as compared to other similar methods, for example the use of transcriptional inhibitors, and they also observe that the method is suitable for detection of plastid transcription. As one reviewer observed, “This method will likely have a large impact on the field of nuclear/organellar RNA metabolism, and the subject is very timely.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00214
Reprogramming of root cells during nitrogen-fixing symbiosis involves dynamic polysome association of coding and noncoding RNAs ($)
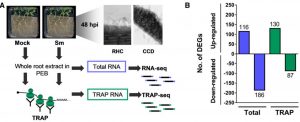 The symbiotic relationship between Rhizobium bacteria and leguminous plants like Medicago results in the development of secondary root organs called nodules. The bacteria housed in the nodule infection zone assimilate atmospheric nitrogen for plant growth. In this paper, Traubenik et al. used (RNA-seq) and translating ribosome affinity purification RNA-seq (TRAP-seq) to determine the translational regulators during nodule organogenesis. Tissue collected from Medicago root 48-hour post-inoculation with rhizobia was used for high throughput sequencing. The correlation between gene expression data from RNA-seq and TRAP-seq indicates the extent of translational regulation during cellular reprogramming in the nodule dedifferentiation process. This sequencing data aided in the identification of several interesting genes. One of these is TAS3, encodes a conserved long noncoding RNA (lncRNA) that produces two alternatively spliced transcripts that are differentially associated with polysomes during bacterial colonization. Another, SKI3, encodes subunit 3 of the SUPERKILLER complex, which participates in mRNA decay, and produces three differentially sized transcripts one of which is translationally upregulated. The functional characterization of SKI3 underlines the significance of this SKI3 protein complex in nodulation as RNAi-induced knock-downs produced significantly fewer nodules. Thus, the TRAP-seq data enhanced the understanding of translational regulation by lncRNA and mRNA decay in Medicago nodule development. (Summary by Suresh Damodaran) Plant Cell 10.1105/tpc.19.00647
The symbiotic relationship between Rhizobium bacteria and leguminous plants like Medicago results in the development of secondary root organs called nodules. The bacteria housed in the nodule infection zone assimilate atmospheric nitrogen for plant growth. In this paper, Traubenik et al. used (RNA-seq) and translating ribosome affinity purification RNA-seq (TRAP-seq) to determine the translational regulators during nodule organogenesis. Tissue collected from Medicago root 48-hour post-inoculation with rhizobia was used for high throughput sequencing. The correlation between gene expression data from RNA-seq and TRAP-seq indicates the extent of translational regulation during cellular reprogramming in the nodule dedifferentiation process. This sequencing data aided in the identification of several interesting genes. One of these is TAS3, encodes a conserved long noncoding RNA (lncRNA) that produces two alternatively spliced transcripts that are differentially associated with polysomes during bacterial colonization. Another, SKI3, encodes subunit 3 of the SUPERKILLER complex, which participates in mRNA decay, and produces three differentially sized transcripts one of which is translationally upregulated. The functional characterization of SKI3 underlines the significance of this SKI3 protein complex in nodulation as RNAi-induced knock-downs produced significantly fewer nodules. Thus, the TRAP-seq data enhanced the understanding of translational regulation by lncRNA and mRNA decay in Medicago nodule development. (Summary by Suresh Damodaran) Plant Cell 10.1105/tpc.19.00647
The pan-genome effector-triggered immunity landscape
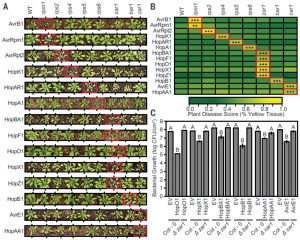 Successful plant pathogens produce a suite of small molecules called effectors that are injected into plant cells and perturb plant defense. As a counter defense, plants have evolved intracellular receptors (called NLRs; nucleotide-binding domain leucine-rich repeat) that detect specific effectors and mount strong effector-triggered immunity (ETI). Compared with an immense diversity of bacteria and their effectors, plants carry a relatively small number of NLRs, but at the same time they are not susceptible to most of the bacterial species. To better understand the role of ETI in determining pathogen host range, Laflamme et al. set out to systematically screen effectors for their ability to trigger ETI in A. thaliana. They created the P. syringae Type III Effector compendium (PsyTEC), from thousands of strains, covering all 70 effector families. By expressing the effectors in P. syringae DC3000 and screening for ETI, they identified 19 ETI-eliciting effector families, which they further characterized using a panel of NLR mutants to identify novel effector-NLR pairs. The results predict that A. thaliana has near-complete immunity to a wide range of P. syringae species, mediated by as few as eight NLRs with a particularly broad contribution from the NLRs ZAR1 and CAR1. This study provides a valuable insight into the role of ETI in constraining bacterial host range and sets the foundation for investigating an even larger set of effector-NLR interactions and effector-effector interactions in the future. (Summary by Tatsuya Nobori) Science 10.1126/science.aax4079
Successful plant pathogens produce a suite of small molecules called effectors that are injected into plant cells and perturb plant defense. As a counter defense, plants have evolved intracellular receptors (called NLRs; nucleotide-binding domain leucine-rich repeat) that detect specific effectors and mount strong effector-triggered immunity (ETI). Compared with an immense diversity of bacteria and their effectors, plants carry a relatively small number of NLRs, but at the same time they are not susceptible to most of the bacterial species. To better understand the role of ETI in determining pathogen host range, Laflamme et al. set out to systematically screen effectors for their ability to trigger ETI in A. thaliana. They created the P. syringae Type III Effector compendium (PsyTEC), from thousands of strains, covering all 70 effector families. By expressing the effectors in P. syringae DC3000 and screening for ETI, they identified 19 ETI-eliciting effector families, which they further characterized using a panel of NLR mutants to identify novel effector-NLR pairs. The results predict that A. thaliana has near-complete immunity to a wide range of P. syringae species, mediated by as few as eight NLRs with a particularly broad contribution from the NLRs ZAR1 and CAR1. This study provides a valuable insight into the role of ETI in constraining bacterial host range and sets the foundation for investigating an even larger set of effector-NLR interactions and effector-effector interactions in the future. (Summary by Tatsuya Nobori) Science 10.1126/science.aax4079
Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis
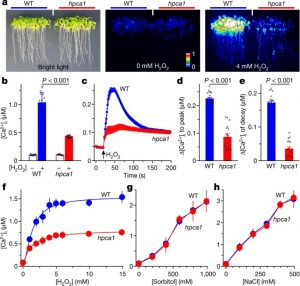 Hydrogen peroxide (H2O2) is an important signal involved in diverse stress responses both intracellularly and extracellularly, but until now it hasn’t been clear how the plant recognizes it. Wu et al. used a genetic screen to search for plants that fail to produce a calcium influx response in the presence of extracellular H2O2 (eH2O2) and identified a cell-surface, leucine-rich-repeat (LRR) receptor kinase that they named HPCA1 (Hydrogen-Peroxide-induced Ca2+ increases). Interestingly, the extracellular domain contains two pairs of Cys residues that appear to form disulfide bonds in the presence of eH2O2, resulting in an activation of the HPCA1 kinase activity. In hpca1 mutants, stomatal closure is impaired as is the calcium response to eH2O2. This study not only shows an interesting new signal receptor, but also, as the authors observe, “can provide potential molecular genetic targets for engineering crops with enhanced fitness and better yield under abiotic and biotic stresses.” (Summary by Mary Williams) Nature 10.1038/s41586-020-2032-3
Hydrogen peroxide (H2O2) is an important signal involved in diverse stress responses both intracellularly and extracellularly, but until now it hasn’t been clear how the plant recognizes it. Wu et al. used a genetic screen to search for plants that fail to produce a calcium influx response in the presence of extracellular H2O2 (eH2O2) and identified a cell-surface, leucine-rich-repeat (LRR) receptor kinase that they named HPCA1 (Hydrogen-Peroxide-induced Ca2+ increases). Interestingly, the extracellular domain contains two pairs of Cys residues that appear to form disulfide bonds in the presence of eH2O2, resulting in an activation of the HPCA1 kinase activity. In hpca1 mutants, stomatal closure is impaired as is the calcium response to eH2O2. This study not only shows an interesting new signal receptor, but also, as the authors observe, “can provide potential molecular genetic targets for engineering crops with enhanced fitness and better yield under abiotic and biotic stresses.” (Summary by Mary Williams) Nature 10.1038/s41586-020-2032-3
Embryonic photosynthesis affects post-germination plant growth
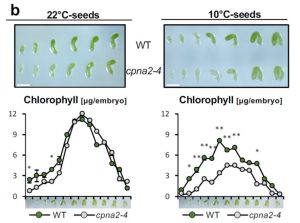 Angiosperm seeds develop within maternal tissues, yet in some species including Arabidopsis the developing embryos carry out photosynthesis. In Arabidopsis, this is transitory, and the embryonic chloroplasts lose chlorophyll and dedifferentiate into eoplasts as the seed matures. Sela et al. set out to investigate whether embryonic photosynthesis is beneficial to the post-germination plant. They identified a cold-induced mutation in a critical chloroplast-localized chaperone. At 10°C, embryos carrying this mutation delayed establishment of a functional photosynthetic apparatus, which allowed the authors to determine the significance of embryonic photosynthesis. The found that perturbing embryonic photosynthesis “leads to profound developmental defects starting from early post-embryonic seedling development and well into the vegetative phase of the plant.” However, as yet it is unclear exactly how this occurs; it appears to not be an affect of altered food stores. It is possible that in wild-type plants the eoplasts that form late in seed development somehow contribute to the plant’s later success, and perturbing this process causes the observed developmental defects. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.20.00043
Angiosperm seeds develop within maternal tissues, yet in some species including Arabidopsis the developing embryos carry out photosynthesis. In Arabidopsis, this is transitory, and the embryonic chloroplasts lose chlorophyll and dedifferentiate into eoplasts as the seed matures. Sela et al. set out to investigate whether embryonic photosynthesis is beneficial to the post-germination plant. They identified a cold-induced mutation in a critical chloroplast-localized chaperone. At 10°C, embryos carrying this mutation delayed establishment of a functional photosynthetic apparatus, which allowed the authors to determine the significance of embryonic photosynthesis. The found that perturbing embryonic photosynthesis “leads to profound developmental defects starting from early post-embryonic seedling development and well into the vegetative phase of the plant.” However, as yet it is unclear exactly how this occurs; it appears to not be an affect of altered food stores. It is possible that in wild-type plants the eoplasts that form late in seed development somehow contribute to the plant’s later success, and perturbing this process causes the observed developmental defects. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.20.00043
HY5 interacts with and recruits HDA9 to negatively regulate autophagy
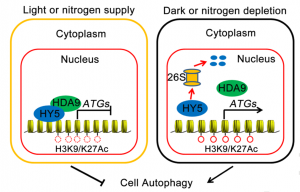 Plants activate autophagy, a cellular recycling process, to recycle useful nutrients or remove harmful items, helping them to withstand harsh conditions. Recently, Yang et al. found that HY5 is a hub connecting the light signaling and autophagy pathways. Under replete conditions (e.g., light and nitrogen-sufficient), the transcription factor HY5 associates with HDA9 and recruits it to autophagy-related genes (ATGs) to repress their expression via histone deacetylation. Under darkness or nitrogen-deficient conditions, HY5 is degraded through the 26s proteasome, HDA9 dissociates from ATGs, and they are derepressed via histone acetylation, thereby activating autophagy. This study demonstrates a previously unknown mechanism of how HY5 associates with histone modification machinery to modulate plant autophagy, and also describes how plants employ different gene expression patterns in response to a fluctuating environment. (Summary by Nanxun Qin) Mol. Plant 10.1016/j.molp.2020.02.011
Plants activate autophagy, a cellular recycling process, to recycle useful nutrients or remove harmful items, helping them to withstand harsh conditions. Recently, Yang et al. found that HY5 is a hub connecting the light signaling and autophagy pathways. Under replete conditions (e.g., light and nitrogen-sufficient), the transcription factor HY5 associates with HDA9 and recruits it to autophagy-related genes (ATGs) to repress their expression via histone deacetylation. Under darkness or nitrogen-deficient conditions, HY5 is degraded through the 26s proteasome, HDA9 dissociates from ATGs, and they are derepressed via histone acetylation, thereby activating autophagy. This study demonstrates a previously unknown mechanism of how HY5 associates with histone modification machinery to modulate plant autophagy, and also describes how plants employ different gene expression patterns in response to a fluctuating environment. (Summary by Nanxun Qin) Mol. Plant 10.1016/j.molp.2020.02.011
Damage-gated immune responses to microbes
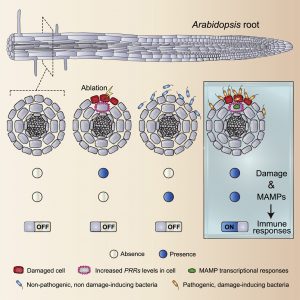 Plants are surrounded by diverse microbes and must avoid mounting an immune response against innocuous microbes, while properly activating defense against invading pathogens. As the initial plant-microbe contacts happen in a limited number of cells, understanding spatially-resolved plant immune responses at the single cell level is important. Zhou et al. used highly sensitive markers to spatially map plant immune activation at cellular resolution in the root. They observed that plant immune responses to flg22, a microbe-derived immune elicitor (microbe-associated molecular pattern; MAMP), are confined to the root cap and transition/elongation zone. Interestingly, flg22 responsiveness could be gained in cells neighboring cells artificially damaged by laser ablation. The authors genetically showed that the flg22 receptor FLS2 is responsible for this damage-gated immune activation. Using double reporter lines of FLS2 and defense marker genes, the authors demonstrated that cellular damage induces neighboring cells to confer flg22 responsiveness in a strictly cell-autonomous manner. These observations were confirmed using actual root-bacteria interaction models. When cells are laser-ablated, non-pathogenic bacteria could trigger immunity in neighboring cells, which are otherwise blind to these bacteria. Moreover, cellular damage caused by root pathogens unlocked the ability of neighboring cells to mount immune responses against the pathogens. Various MAMP receptors were shown to be induced when neighboring cells are damaged. This study deepened our understanding of how plant roots deal with heterogeneous and microbe-rich environments. (Summary by Tatsuya Nobori) Cell 10.1016/j.cell.2020.01.013
Plants are surrounded by diverse microbes and must avoid mounting an immune response against innocuous microbes, while properly activating defense against invading pathogens. As the initial plant-microbe contacts happen in a limited number of cells, understanding spatially-resolved plant immune responses at the single cell level is important. Zhou et al. used highly sensitive markers to spatially map plant immune activation at cellular resolution in the root. They observed that plant immune responses to flg22, a microbe-derived immune elicitor (microbe-associated molecular pattern; MAMP), are confined to the root cap and transition/elongation zone. Interestingly, flg22 responsiveness could be gained in cells neighboring cells artificially damaged by laser ablation. The authors genetically showed that the flg22 receptor FLS2 is responsible for this damage-gated immune activation. Using double reporter lines of FLS2 and defense marker genes, the authors demonstrated that cellular damage induces neighboring cells to confer flg22 responsiveness in a strictly cell-autonomous manner. These observations were confirmed using actual root-bacteria interaction models. When cells are laser-ablated, non-pathogenic bacteria could trigger immunity in neighboring cells, which are otherwise blind to these bacteria. Moreover, cellular damage caused by root pathogens unlocked the ability of neighboring cells to mount immune responses against the pathogens. Various MAMP receptors were shown to be induced when neighboring cells are damaged. This study deepened our understanding of how plant roots deal with heterogeneous and microbe-rich environments. (Summary by Tatsuya Nobori) Cell 10.1016/j.cell.2020.01.013
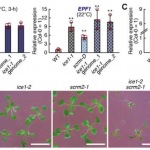
Does ICE1 participate in cold responses?
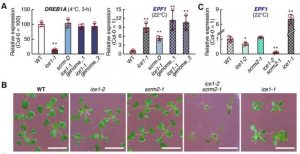 For many years, the Arabidopsis transgenic allele ice1-1 was used to demonstrate the role of the transcription factor ICE1 in freezing and cold tolerance. This gene was first identified in a screening strategy using a luciferase reporter for DREB1A expression, another TF involved in downstream signaling. New results from Kikodore et al. showed that the transgenic reporter induced DNA methylation in the DREB1a locus, suggesting that the role of ICE1 should be re-assessed. In addition, an alternative loss-of-function allele of ICE1 and its homolog SCREAM2 showed a minor or no contribution to cold tolerance and DREB1a expression. These results contrast with many studies using ice1-1 but also alternative strategies to analyze the role of ICE1.
For many years, the Arabidopsis transgenic allele ice1-1 was used to demonstrate the role of the transcription factor ICE1 in freezing and cold tolerance. This gene was first identified in a screening strategy using a luciferase reporter for DREB1A expression, another TF involved in downstream signaling. New results from Kikodore et al. showed that the transgenic reporter induced DNA methylation in the DREB1a locus, suggesting that the role of ICE1 should be re-assessed. In addition, an alternative loss-of-function allele of ICE1 and its homolog SCREAM2 showed a minor or no contribution to cold tolerance and DREB1a expression. These results contrast with many studies using ice1-1 but also alternative strategies to analyze the role of ICE1.
As a response to this paper, Tang et al. discuss the evidence in favor of ICE1 having role in cold stress and present new data using ChiP-seq that suggest that ICE1 directly binds the DREB1A locus and also other genes associated with cold/freezing tolerance.
It is still a matter of controversy whether ICE1 contributes to cold tolerance and how it is related with this other role in stomatal development. In a commentary, Thomashow and Torii discuss these and other aspects.
In summary, these papers put on the debate the limitations of molecular techniques such as reverse genetic screening for functional biology and the importance of evolutionary aspects and using alternative sources of evidence. (Summary by Facundo Romani)
Kikodore et al. https://doi.org/10.1105/tpc.19.00532
Tang et al. https://doi.org/10.1111/jipb.12918
Thomashow and Torii https://doi.org/10.1105/tpc.20.00124