Plant Science Research Weekly: December 16, 2022
Review: The epigenetic control of the transposable element life cycle in plant genomes and beyond
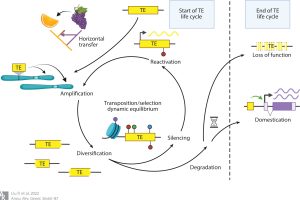 What a wonderful review! As the title indicates, this review by Liu et al. looks at transposons, considering their “life cycle” – from introduction into a naïve genome, through amplification, diversification, silencing, reactivation, and ultimately domestication or loss-of-function. Besides this thorough-but-readable walk through the transposon life cycle, the review also takes a pan-organismal look at transposons and their regulators, with strong representation from the plant kingdom. There’s also a discussion about how various transposon families got their names (interesting!) and efforts to both standardize them and eliminate names with derogatory origins. The final two sections were to me the most interesting. First, how TEs are “tamed” by reuse and domestication; TEs can be repurposed by incorporation into new genes, new regulatory elements, or telomeres. And finally, human interactions with TEs, which have advanced our understanding of evolution and adaptation; TEs were responsible for Mendel’s wrinkled peas, and the emergence of dark moths during the sooty coal-fueled industrial revolution. Furthermore, TEs have contributed to the genetic diversity in crop breeding and, in news to me, the diversification of dog breeds. Woof! (Summary by Mary Williams @PlantTeaching) Annu. Rev. Genetics 10.1146/annurev-genet-072920-015534
What a wonderful review! As the title indicates, this review by Liu et al. looks at transposons, considering their “life cycle” – from introduction into a naïve genome, through amplification, diversification, silencing, reactivation, and ultimately domestication or loss-of-function. Besides this thorough-but-readable walk through the transposon life cycle, the review also takes a pan-organismal look at transposons and their regulators, with strong representation from the plant kingdom. There’s also a discussion about how various transposon families got their names (interesting!) and efforts to both standardize them and eliminate names with derogatory origins. The final two sections were to me the most interesting. First, how TEs are “tamed” by reuse and domestication; TEs can be repurposed by incorporation into new genes, new regulatory elements, or telomeres. And finally, human interactions with TEs, which have advanced our understanding of evolution and adaptation; TEs were responsible for Mendel’s wrinkled peas, and the emergence of dark moths during the sooty coal-fueled industrial revolution. Furthermore, TEs have contributed to the genetic diversity in crop breeding and, in news to me, the diversification of dog breeds. Woof! (Summary by Mary Williams @PlantTeaching) Annu. Rev. Genetics 10.1146/annurev-genet-072920-015534
Review: Beyond transcription: compelling open questions in plant RNA biology
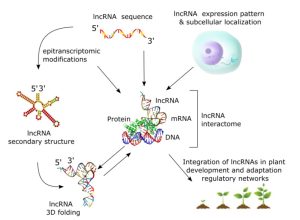 The May 2023 issue of The Plant Cell has a focus on RNA biology, and some of the articles for this issue are online now. I particularly enjoyed this multi-author perspective review by Manavella et al., in which 12 RNA biology groups from across the globe share the questions that they find most compelling in their field today. Their short vignettes collectively address “RNA alternative splicing; RNA dynamics; RNA translation; RNA structures; R-loops; epitranscriptomics; long noncoding RNAs; small RNA production and their functions in crops; small RNAs during gametogenesis and in cross-kingdom RNA interference; and RNA-directed DNA methylation,” so there really is something for everyone here. Helpfully, each also has a short update of the state-of-the art of the discipline, making this an excellent resources to catch up and look ahead. There are few fields that have developed so quickly and recently as RNA biology, which surges ahead with each new methodological advancement, and which touches so pervasively on all facets of the plant life cycle, literally from birth to death. I hope you find time to read this information-packed and insightful review article. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koac346
The May 2023 issue of The Plant Cell has a focus on RNA biology, and some of the articles for this issue are online now. I particularly enjoyed this multi-author perspective review by Manavella et al., in which 12 RNA biology groups from across the globe share the questions that they find most compelling in their field today. Their short vignettes collectively address “RNA alternative splicing; RNA dynamics; RNA translation; RNA structures; R-loops; epitranscriptomics; long noncoding RNAs; small RNA production and their functions in crops; small RNAs during gametogenesis and in cross-kingdom RNA interference; and RNA-directed DNA methylation,” so there really is something for everyone here. Helpfully, each also has a short update of the state-of-the art of the discipline, making this an excellent resources to catch up and look ahead. There are few fields that have developed so quickly and recently as RNA biology, which surges ahead with each new methodological advancement, and which touches so pervasively on all facets of the plant life cycle, literally from birth to death. I hope you find time to read this information-packed and insightful review article. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koac346
A family of methyl esterases converts methyl salicylate to salicylic acid in ripening tomato fruit
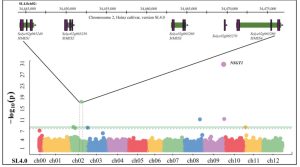 This is an interesting paper that grabbed my attention from the very first line: “Tomatoes are the second most valuable crop in the USA”. Of course, a key trait in these important fruits is their flavor, something the Harry Klee team at the University of Florida have been studying for years. Here, Frick et al. investigated the conversion of methyl salicylate (negatively correlated with good flavor) to salicylate in tomato. They used a GWAS approach to identify a region associated with methyl salicylate abundance that includes four genes encoding methyl esterases, which they named SlMES1-4 (I misread this a SIMES to rhyme with limes, but it the second letter is a lower-case l, for Solanum lycopersicum MES). Several lines of evidence including CRISPR editing implicated SlMES1 as having a major role in the demethylation of methyl salicylate in tomato fruit, with the other three genes having smaller effects. Heterologous expression confirmed SlMES1 as having methyl esterase activity against methyl salicylate. This work provides a new tool for breeders to decrease a negative component of flavor, but it is also interesting because methyl salicylate is an important defense compound in plants, related to and more mobile than salicylic acid (SA). Can genetics tease apart the the flavor-defense trade off? (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiac509
This is an interesting paper that grabbed my attention from the very first line: “Tomatoes are the second most valuable crop in the USA”. Of course, a key trait in these important fruits is their flavor, something the Harry Klee team at the University of Florida have been studying for years. Here, Frick et al. investigated the conversion of methyl salicylate (negatively correlated with good flavor) to salicylate in tomato. They used a GWAS approach to identify a region associated with methyl salicylate abundance that includes four genes encoding methyl esterases, which they named SlMES1-4 (I misread this a SIMES to rhyme with limes, but it the second letter is a lower-case l, for Solanum lycopersicum MES). Several lines of evidence including CRISPR editing implicated SlMES1 as having a major role in the demethylation of methyl salicylate in tomato fruit, with the other three genes having smaller effects. Heterologous expression confirmed SlMES1 as having methyl esterase activity against methyl salicylate. This work provides a new tool for breeders to decrease a negative component of flavor, but it is also interesting because methyl salicylate is an important defense compound in plants, related to and more mobile than salicylic acid (SA). Can genetics tease apart the the flavor-defense trade off? (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiac509
SYNTAXIN OF PLANTS81 regulates root meristem activity and stem cell niche maintenance via ROS signaling
 Lately, reactive oxygen species (ROS) have been recognized as signaling molecules that regulate plant cellular proliferation and differentiation in many areas of the plant, including root tips. Chloroplasts, peroxisomes, and mitochondria are the main cellular compartments for ROS generation in cells. SYNTAXIN OF PLANTS81 in Arabidopsis thaliana (AtSYP81), a SNARE protein, has been suggested to be involved in ER-Golgi trafficking in the plant secretory pathway, but its role is still unclear. In this article, Wang et al. discovered that AtSYP81 regulates ROS homeostasis by controlling root apical meristem activity and the maintenance of root stem cell niche. Through GUS staining and fluorescently-tagged proteins, the authors found that atsyp81 mutants showed fewer peroxisomes in root meristem cells, and ROS levels were decreased in atsyp81 roots.In addition, transcriptomic analysis between atsyp81 and AtSYP81 overexpression line showed that class III peroxidases were differentially expressed. The authors suggest a mechanism involving PLETHORA 1 and 2 (PLT1/2) transcription factors, downstream of AtSYP81 and through auxin signaling. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Physiol. 10.1093/plphys/kiac530
Lately, reactive oxygen species (ROS) have been recognized as signaling molecules that regulate plant cellular proliferation and differentiation in many areas of the plant, including root tips. Chloroplasts, peroxisomes, and mitochondria are the main cellular compartments for ROS generation in cells. SYNTAXIN OF PLANTS81 in Arabidopsis thaliana (AtSYP81), a SNARE protein, has been suggested to be involved in ER-Golgi trafficking in the plant secretory pathway, but its role is still unclear. In this article, Wang et al. discovered that AtSYP81 regulates ROS homeostasis by controlling root apical meristem activity and the maintenance of root stem cell niche. Through GUS staining and fluorescently-tagged proteins, the authors found that atsyp81 mutants showed fewer peroxisomes in root meristem cells, and ROS levels were decreased in atsyp81 roots.In addition, transcriptomic analysis between atsyp81 and AtSYP81 overexpression line showed that class III peroxidases were differentially expressed. The authors suggest a mechanism involving PLETHORA 1 and 2 (PLT1/2) transcription factors, downstream of AtSYP81 and through auxin signaling. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Physiol. 10.1093/plphys/kiac530
Genome-wide dissection of changes in maize root system architecture during modern breeding
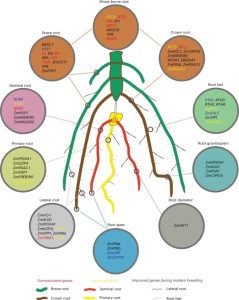 Root system architecture (RSA) is a complex trait that directly or indirectly influences most aspects of plant survival, largely through its effects on nutrient and water uptake as well as support and anchoring. Its hidden nature has made it harder to study than the architecture of the shoot, and led to many innovative methods. Here, Ren et al. applied an association study to identify genes affecting RSA, as characterized by a “shovelomics” approach involving 40 x 30 cm samples dug from the base of the shoot. They assessed 380 inbred accessions of maize, focusing on adult field-grown plants, and measuring eight root traits from approximately 15,000 individuals across four conditions. They made a lot of observations, including documenting changes in RSA across time by looking at accessions released in China from the 1960s to 2010s; during this period, root angles became steeper and root widths narrower. This very large study identified a very large number of genes-of-interest, many of which showed clear signs of selection during modern Chinese maize breeding programs. Functional studies of a few previously uncharacterized genes revealed strong effects on RSA through effects on auxin signaling. For those who dig roots, there’s plenty more data to enjoy! (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-022-01274-z
Root system architecture (RSA) is a complex trait that directly or indirectly influences most aspects of plant survival, largely through its effects on nutrient and water uptake as well as support and anchoring. Its hidden nature has made it harder to study than the architecture of the shoot, and led to many innovative methods. Here, Ren et al. applied an association study to identify genes affecting RSA, as characterized by a “shovelomics” approach involving 40 x 30 cm samples dug from the base of the shoot. They assessed 380 inbred accessions of maize, focusing on adult field-grown plants, and measuring eight root traits from approximately 15,000 individuals across four conditions. They made a lot of observations, including documenting changes in RSA across time by looking at accessions released in China from the 1960s to 2010s; during this period, root angles became steeper and root widths narrower. This very large study identified a very large number of genes-of-interest, many of which showed clear signs of selection during modern Chinese maize breeding programs. Functional studies of a few previously uncharacterized genes revealed strong effects on RSA through effects on auxin signaling. For those who dig roots, there’s plenty more data to enjoy! (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-022-01274-z
Best wishes for a healthy and happy new year from all of us at Plant Science Research Weekly! We’ll be back in 2023 with more exciting plant science. Don’t forget you can subscribe to this series at https://plantae.org/research/wwrtw/.