Genome-wide dissection of maize root system architecture
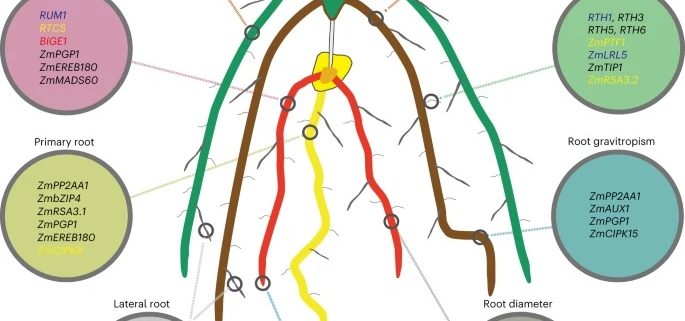
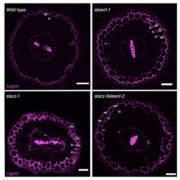
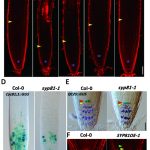
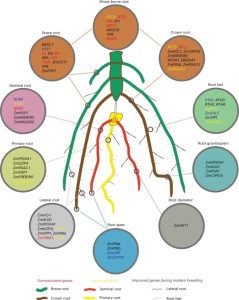 Root system architecture (RSA) is a complex trait that directly or indirectly influences most aspects of plant survival, largely through its effects on nutrient and water uptake as well as support and anchoring. Its hidden nature has made it harder to study than the architecture of the shoot, and led to many innovative methods. Here, Ren et al. applied an association study to identify genes affecting RSA, as characterized by a “shovelomics” approach involving 40 x 30 cm samples dug from the base of the shoot. They assessed 380 inbred accessions of maize, focusing on adult field-grown plants, and measuring eight root traits from approximately 15,000 individuals across four conditions. They made a lot of observations, including documenting changes in RSA across time by looking at accessions released in China from the 1960s to 2010s; during this period, root angles became steeper and root widths narrower. This very large study identified a very large number of genes-of-interest, many of which showed clear signs of selection during modern Chinese maize breeding programs. Functional studies of a few previously uncharacterized genes revealed strong effects on RSA through effects on auxin signaling. For those who dig roots, there’s plenty more data to enjoy! (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-022-01274-z
Root system architecture (RSA) is a complex trait that directly or indirectly influences most aspects of plant survival, largely through its effects on nutrient and water uptake as well as support and anchoring. Its hidden nature has made it harder to study than the architecture of the shoot, and led to many innovative methods. Here, Ren et al. applied an association study to identify genes affecting RSA, as characterized by a “shovelomics” approach involving 40 x 30 cm samples dug from the base of the shoot. They assessed 380 inbred accessions of maize, focusing on adult field-grown plants, and measuring eight root traits from approximately 15,000 individuals across four conditions. They made a lot of observations, including documenting changes in RSA across time by looking at accessions released in China from the 1960s to 2010s; during this period, root angles became steeper and root widths narrower. This very large study identified a very large number of genes-of-interest, many of which showed clear signs of selection during modern Chinese maize breeding programs. Functional studies of a few previously uncharacterized genes revealed strong effects on RSA through effects on auxin signaling. For those who dig roots, there’s plenty more data to enjoy! (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-022-01274-z